Paper:
Control of Osmotic-Engine-Driven Liposomes Using Biological Nanopores
Hinata Shibuya, Shun Okada, and Kan Shoji

Department of Mechanical Engineering, Nagaoka University of Technology
1603-1 Kamitomioka, Nagaoka, Niigata 940-2188, Japan
Liposome-based molecular robots that molecular systems are integrated into a giant liposome have been proposed; they are expected to be applied in the fields of medicine, environmental science, food science, and energy science. However, the performance of these molecular robotic components, including intelligence, sensors, and actuators, still hinders their practical use. In particular, the actuators used in the molecular robots, such as molecular motors, do not provide sufficient performance to move the giant liposomes. Hence, we propose an osmotic-engine-driven liposome and demonstrate the migration of liposomes in a microfluidic channel by applying a salt concentration difference between the front and rear of the liposome. Although the migration mechanism is simple and has the potential to provide sufficient mobility performance, control techniques for the movement speed and on/off switching are not established. Herein, we describe a speed control method of osmotic-engine-driven liposomes using pore-forming membrane proteins. In this study, we evaluated the effect of reconstituted α-hemolysin (αHL) nanopores on the water permeability through lipid bilayers. Thereafter, we demonstrated the change in displacement speeds of liposomes with and without nanopores. We expect the speed control method using nanopores to be applied to the liposome-based molecular robots.
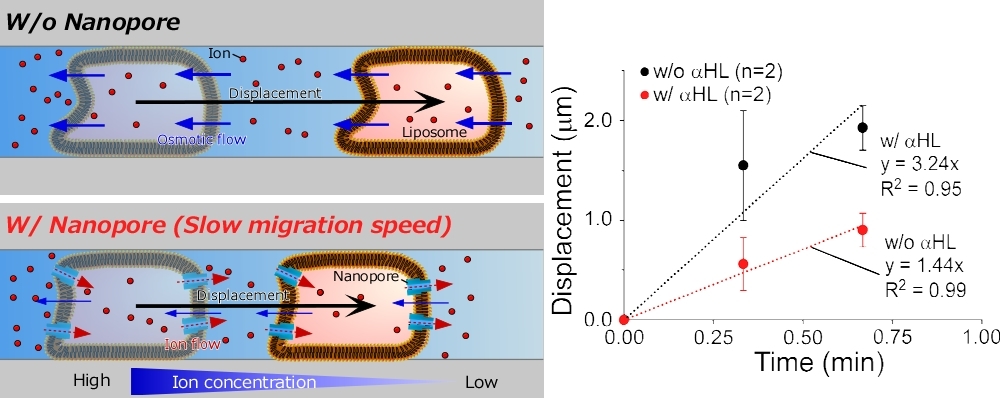
Liposome migration with and without biological nanopores
- [1] S. Murata, A. Konagaya, S. Kobayashi, H. Saito, and M. Hagiya, “Molecular Robotics: A New Paradigm for Artifacts,” New Gener. Comput., Vol.31, No.1, pp. 27-45, 2013. https://doi.org/10.1007/s00354-012-0121-z
- [2] M. Hagiya, A. Konagaya, S. Kobayashi, H. Saito, and S. Murata, “Molecular Robots with Sensors and Intelligence,” Acc. Chem. Res., Vol.47, No.6, pp. 1681-1690, 2014. https://doi.org/10.1021/ar400318d
- [3] R. Kawano, “Synthetic Ion Channels and DNA Logic Gates as Components of Molecular Robots,” ChemPhysChem, Vol.19, No.4, pp. 359-366, 2018. https://doi.org/10.1002/cphc.201700982
- [4] K. Shoji and R. Kawano, “Recent Advances in Liposome-Based Molecular Robots,” Micromachines, Vol.11, No.9, Article No.788, 2020. https://doi.org/10.3390/mi11090788
- [5] S. Murata, T. Toyota, S. M. Nomura, T. Nakakuki, and A. Kuzuya, “Molecular Cybernetics: Challenges Toward Cellular Chemical Artificial Intelligence,” Advanced Functional Materials, Vol.32, No.37, Article No.2270207, 2022. https://doi.org/10.1002/adfm.202201866
- [6] K. Akashi, H. Miyata, H. Itoh, and K. Kinosita, Jr., “Preparation of giant liposomes in physiological conditions and their characterization under an optical microscope,” Biophysical J., Vol.71, No.6, pp. 3242-3250, 1996. https://doi.org/10.1016/S0006-3495(96)79517-6
- [7] A. Moscho, O. Orwar, D. T. Chiu, B. P. Modi, and R. N. Zare, “Rapid preparation of giant unilamellar vesicles,” Proc. of the National Academy of Sciences, Vol.93, No.21, pp. 11443-11447, 1996. https://doi.org/10.1073/pnas.93.21.11443
- [8] C. Kurokawa et al., “DNA cytoskeleton for stabilizing artificial cells,” Proc. of the National Academy of Sciences, Vol.114, No.28, pp. 7228-7233, 2017. https://doi.org/10.1073/pnas.1702208114
- [9] T. Tohgasaki et al., “A Photocaged DNA Nanocapsule for Controlled Unlocking and Opening inside the Cell,” Bioconjugate Chem., Vol.30, No.7, pp. 1860-1863, 2019. https://doi.org/10.1021/acs.bioconjchem.9b00040
- [10] M. Hagiya et al., “On DNA-Based Gellular Automata,” O. H. Ibarra, L. Kari, and S. Kopecki (Eds.), “Lecture Notes in Computer Science: 13th Int. Conf. on Unconventional Computation and Natural Computation,” pp. 177-189, Springer, 2014. https://doi.org/10.1007/978-3-319-08123-6_15
- [11] K. Abe, I. Kawamata, S. M. Nomura, and S. Murata, “Programmable reactions and diffusion using DNA for pattern formation in hydrogel medium,” Mol. Syst. Des. Eng., Vol.4, No.3, pp. 639-643, 2019. https://doi.org/10.1039/C9ME00004F
- [12] H. Bermudez, A. K. Brannan, D. A. Hammer, F. S. Bates, and D. E. Discher, “Molecular Weight Dependence of Polymersome Membrane Structure, Elasticity, and Stability,” Macromolecules, Vol.35, No.21, pp. 8203-8208, 2002. https://doi.org/10.1021/ma020669l
- [13] M. Honda, K. Takiguchi, S. Ishikawa, and H. Hotani, “Morphogenesis of liposomes encapsulating actin depends on the type of actin-crosslinking,” J. of Molecular Biology, Vol.287, No.2, pp. 293-300, 1999. https://doi.org/10.1006/jmbi.1999.2592
- [14] H. Hotani, F. Nomura, and Y. Suzuki, “Giant liposomes: From membrane dynamics to cell morphogenesis,” Current Opinion in Colloid & Interface Science, Vol.4, No.5, pp. 358-368, 1999. https://doi.org/10.1016/S1359-0294(99)90021-3
- [15] M. Hayashi, M. Nishiyama, Y. Kazayama, T. Toyota, Y. Harada, and K. Takiguchi, “Reversible Morphological Control of Tubulin-Encapsulating Giant Liposomes by Hydrostatic Pressure,” Langmuir, Vol.32, No.15, pp. 3794-3802, 2016. https://doi.org/10.1021/acs.langmuir.6b00799
- [16] S. Tanaka, K. Takiguchi, and M. Hayashi, “Repetitive stretching of giant liposomes utilizing the nematic alignment of confined actin,” Commun. Phys., Vol.1, Article No.18, 2018. https://doi.org/10.1038/s42005-018-0019-2
- [17] Y. Sato, Y. Hiratsuka, I. Kawamata, S. Murata, and S. M. Nomura, “Micrometer-sized molecular robot changes its shape in response to signal molecules,” Science Robotics, Vol.2, No.4, Article No.eaal3735, 2017. https://doi.org/10.1126/scirobotics.aal3735
- [18] M. Nakajima et al., “Quantitative Evaluation of Injected Molecules into Phospholipid-Coated Microdroplets for In situ Biological Reactions,” J. Robot. Mechatron., Vol.22, No.5, pp. 651-658, 2010. https://doi.org/10.20965/jrm.2010.p0651
- [19] K. Shoji and R. Kawano, “Osmotic-engine-driven liposomes in microfluidic channels,” Lab on a Chip, Vol.19, No.20, pp. 3472-3480, 2019. https://doi.org/10.1039/C9LC00788A
- [20] K. M. Stroka et al., “Water Permeation Drives Tumor Cell Migration in Confined Microenvironments,” Cell, Vol.157, No.3, pp. 611-623, 2014. https://doi.org/10.1016/j.cell.2014.02.052
- [21] K. Funakoshi, H. Suzuki, and S. Takeuchi, “Lipid Bilayer Formation by Contacting Monolayers in a Microfluidic Device for Membrane Protein Analysis,” Anal. Chem., Vol.78, No.24, pp. 8169-8174, 2006. https://doi.org/10.1021/ac0613479
- [22] R. Kawano et al., “Automated Parallel Recordings of Topologically Identified Single Ion Channels,” Sci. Rep., Vol.3, Article No.1995, 2013. https://doi.org/10.1038/srep01995
- [23] K. Shoji, R. Kawano, and R. J. White, “Recessed Ag/AgCl Microelectrode-Supported Lipid Bilayer for Nanopore Sensing,” Anal. Chem., Vol.92, No.15, pp. 10856-10862, 2020. https://doi.org/10.1021/acs.analchem.0c02720
- [24] S. Pautot, B. J. Frisken, and D. A. Weitz, “Production of Unilamellar Vesicles Using an Inverted Emulsion,” Langmuir, Vol.19, No.7, pp. 2870-2879, 2003. https://doi.org/10.1021/la026100v
- [25] N. N. Deng, M. Yelleswarapu, and W. T. S. Huck, “Monodisperse Uni- and Multicompartment Liposomes,” J. Am. Chem. Soc., Vol.138, No.24, pp. 7584-7591, 2016. https://doi.org/10.1021/jacs.6b02107
- [26] F. A. Edwards, A. J. Gibb, and D. Colquhoun, “ATP receptor-mediated synaptic currents in the central nervous system,” Nature, Vol.359, No.6391, pp. 144-147, 1992. https://doi.org/10.1038/359144a0
- [27] P. M. Arnott and S. Howorka, “A Temperature-Gated Nanovalve Self-Assembled from DNA to Control Molecular Transport Across Membranes,” ACS Nano, Vol.13, No.3, pp. 3334-3340, 2019. https://doi.org/10.1021/acsnaNo.8b09200
- [28] J. R. Burns, A. Seifert, N. Fertig, and S. Howorka, “A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane,” Nature Nanotech., Vol.11, No.2, pp. 152-156, 2016. https://doi.org/10.1038/nnaNo.2015.279
- [29] C. Lanphere, P. M. Arnott, S. F. Jones, K. Korlova, and S. Howorka, “A Biomimetic DNA-Based Membrane Gate for Protein-Controlled Transport of Cytotoxic Drugs,” Angewandte Chemie Int. Edition, Vol.60, No.4, pp. 1903-1908, 2021. https://doi.org/10.1002/anie.202011583
- [30] D. Offenbartl-Stiegert, A. Rottensteiner, A. Dorey, and S. Howorka, “A Light-Triggered Synthetic Nanopore for Controlling Molecular Transport Across Biological Membranes,” Angewandte Chemie Int. Edition, Vol.61, No.52, Article No.e202210886, 2022. https://doi.org/10.1002/anie.202210886
 This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.
This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.