Paper:
Varietal Classification of Lactuca Sativa Seeds Using an Adaptive Neuro-Fuzzy Inference System Based on Morphological Phenes
Christan Hail R. Mendigoria*,†, Heinrick L. Aquino*, Oliver John Y. Alajas*, Ronnie S. Concepcion II*, Elmer P. Dadios**, Edwin Sybingco*, Argel A. Bandala*, and Ryan Rhay P. Vicerra**
*Electronics and Communications Engineering Department, De La Salle University
2401 Taft Ave, Malate, Manila 1004, Philippines
**Manufacturing Engineering and Management Department, De La Salle University
2401 Taft Ave, Malate, Manila 1004, Philippines
†Corresponding author
Seed varieties are often differentiated via the manual and subjective classification of their external textural, spectral, and morphological biosignatures. This traditional method of manually inspecting seeds is inefficient and unreliable for seed phenotyping. The application of computer vision is an ideal solution allied with computational intelligence. This study used Lactuca sativa seed variants, which are commercially known as grand rapid, Chinese loose-leaf, and iceberg (which serves as noise data for extended model evaluation), in determining their corresponding classifications based on the extended morphological phenes using computational intelligence. Red-green-blue (RGB) imaging was employed for individual kernels. Extended morphological phenes, that is, solidity, roundness, compactness, and shape factors, were computed based on seed architectural traits and used as predictors to discriminate among the three cultivars. The suitability of ANFIS, NB, and CT was explored using a limited dataset. A mean accuracy of 100% was manifested in ANFIS; thus, it was proved to be the most reliable model.
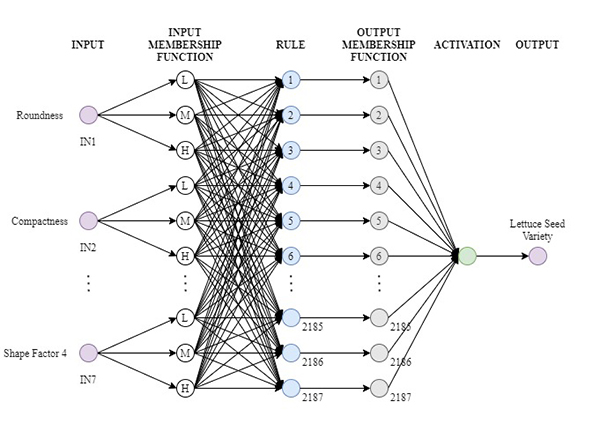
Lettuce seed classification (FIS) model
- [1] R. Concepcion II et al., “Variety Classification of Lactuca Sativa Seeds Using Single-Kernel RGB Images and Spectro- Textural-Morphological Feature-Based Machine Learning,” IEEE Int. Conf. on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment and Management, 2020.
- [2] D. P. Van Hoai et al., “A comparative study of rice variety classification based on deep learning and hand-crafted features,” ECTI Trans. Comput. Inf. Technol., Vol.14, No.1, pp. 1-10, 2020.
- [3] J. Alejandrino et al., “Visual classification of lettuce growth stage based on morphological attributes using unsupervised machine learning models,” 2020 IEEE Region 10 Conf. (TENCON), pp. 438-443, 2020.
- [4] S. C. Lauguico et al., “Implementation of Inverse Kinematics for Crop-Harvesting Robotic Arm in Vertical Farming,” Proc. of the IEEE 2019 9th Int. Conf. on Cybernetics and Intelligent Systems (CIS) and Robotics, Automation and Mechatronics (RAM), 2019.
- [5] S. C. Lauguico et al., “Lettuce life stage classification from texture attributes using machine learning estimators and feature selection processes,” Int. J. Adv. Intell. Informatics, Vol.6, No.2, pp. 173-184, 2020.
- [6] R. G. De Luna, E. P. Dadios, and A. A. Bandala, “Automated Image Capturing System for Deep Learning-based Tomato Plant Leaf Disease Detection and Recognition,” 2018 IEEE Region 10 Conf. (TENCON 2018), pp. 1414-1419, 2018.
- [7] R. Concepcion II et al., “Lettuce growth stage identification based on phytomorphological variations using coupled color superpixels and multifold watershed transformation,” Int. J. Adv. Intell. Informatics, Vol.6, No.3, pp. 261-277, 2020.
- [8] V. d. J. M. Bianchini et al., “Multispectral and X-ray images for characterization of Jatropha curcas L. seed quality,” Plant Methods, Vol.17, Article No.9, 2021.
- [9] Q. Zhou et al., “Non-destructive discrimination of the variety of sweet maize seeds based on hyperspectral image coupled with wavelength selection algorithm,” Infrared Phys. Technol., Vol.109, Article No.103418, 2020.
- [10] R. Moscetti et al., “Pine nut species recognition using NIR spectroscopy and image analysis,” J. Food Eng., Vol.292, Article No.110357, 2021.
- [11] J. Liu et al., “A modified feature fusion method for distinguishing seed strains using hyperspectral data,” Int. J. Food Eng., Vol.16, No.7, pp. 1-12, 2020.
- [12] F. Kurtulmuş, “Identification of sunflower seeds with deep convolutional neural networks,” J. Food Meas. Charact., Vol.15, No.2, pp. 1024-1033, 2021.
- [13] X. Hong, S. Guan, K. L. Man, and P. W. H. Wong, “A Convolution Neural Network-Based Seed Classification System,” Symmetry, Vol.12, Article No.2018, 2020.
- [14] J. Yasmin et al., “Improvement in Purity of Healthy Tomato Seeds Using an Image-Based One-Class Classification Method,” Sensors, Vol.20, Article No.2690, 2020.
- [15] S. D. Fabiyi et al., “Varietal Classification of Rice Seeds Using RGB and Hyperspectral Images,” IEEE Access, Vol.8, pp. 22493-22505, 2020.
- [16] J. A. A. Garcia, E. R. Arboleda, and E. M. Galas, “Identification of visually similar vegetable seeds using image processing and fuzzy logic,” Int. J. Sci. Technol. Res., Vol.9, No.2, pp. 4925-4928, 2020.
- [17] P. Singh et al., “Classification of wheat seeds using image processing and fuzzy clustered random forest,” Int. J. Agric. Resour. Gov. Ecol., Vol.16, No.2, pp. 123-156, 2020.
- [18] V. Goel et al., “Specific Color Detection in Images using RGB Modelling in MATLAB,” Int. J. of Computer Applications, Vol.161, No.8, pp. 38-42, doi: 10.5120/ijca2017913254, 2017.
- [19] R. Concepcion II et al., “Genetic Algorithm-Based Visible Band Tetrahedron Greenness Index Modeling for Lettuce Biophysical Signature Estimation,” 2020 IEEE Region 10 Conf. (TENCON), 2020.
- [20] V. J. Almero et al., “An Aquaculture-Based Binary Classifier for Fish Detection using Multilayer Artificial Neural Network,” 2019 IEEE 11th Int. Conf. on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment, and Management (HNICEM), 2019.
 This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.
This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.