Research Paper:
Can Sleep Apnea Be Detected from Human Pulse Waveform with Laplace Noise?
Itaru Kaneko*
 , Le Trieu Phong**
, Le Trieu Phong**
 , Keita Emura**
, Keita Emura**
 , and Emi Yuda*,†
, and Emi Yuda*,†

*Graduate School of Information Sciences, Tohoku University
6-3-09 Aramaki-Aza-Aoba, Aoba-ku, Sendai, Miyagi 980-8579, Japan
†Corresponding author
**National Institute of Information and Communications Technology (NICT)
4-2-1 Nukui-Kitamachi, Koganei, Tokyo 184-8795, Japan
Differential privacy is a powerful technique that protects the privacy of individuals in a dataset by adding controlled randomness. With the increasing developments in smart sensors, the use of various biometric database is expanding. If privacy protections coexist with advanced use of the biometric database, wider utilization is expected. One of the promising approaches is to apply differential privacy to biometric information, which is attracting attention in use cases such as Google. By adding Laplace noise to biometric information, differential privacy can be added. Our aim is to focus on peak to peak interval of electrocardiogram. It is useful bio-information because it is possible to know not only heart disease but also various physical conditions such as exercise amount, activity amount, fatigue, sleep based on it. In this study, we demonstrated that differential privacy can be applied to obtain the sleep apnea index from PPIs with Laplace noise. The observed correlations were 0.96 to 0.99 for the corresponding PPIs with Laplace noise.
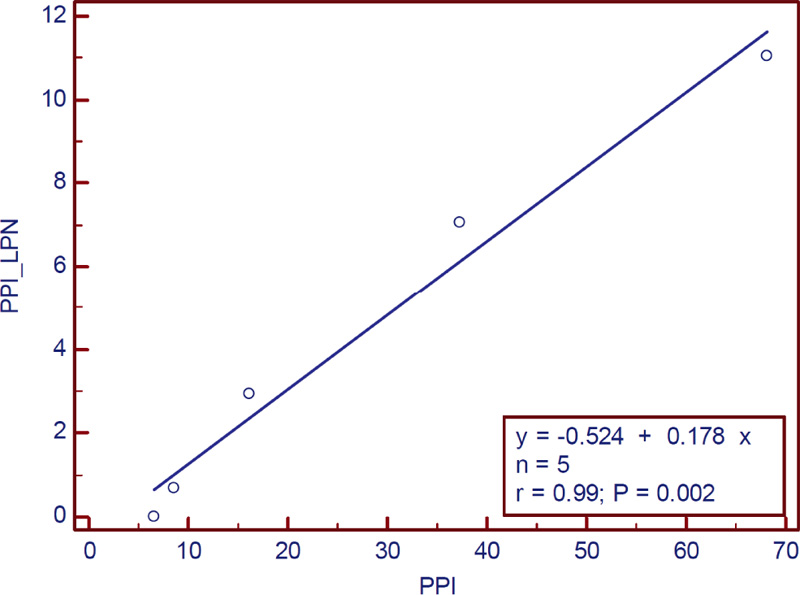
Correlation between CVHR calculated from PPI with Laplace noise and PPI
- [1] K. Jensen-Urstad, N. Storck, F. Bouvier, M. Ericson, L. E. Lindbland, and M. Jensen-Urstad, “Heart rate variability in healthy subjects is related to age and gender,” Acta Physiol. Scand., Vol.160, Issue 3, pp. 235-241, 1997. https://doi.org/10.1046/j.1365-201X.1997.00142.x
- [2] M. W. Agelink, R. Malessa, B. Baumann, T. Majewski, F. Akila, T. Zeit, and D. Ziegler, “Standardized tests of heart rate variability: Normal ranges obtained from 309 healthy humans, and effects of age, gender, and heart rate,” Clin. Auton. Res., Vol.11, No.2, pp. 99-108, 2001. https://doi.org/10.1007/BF02322053
- [3] T. Yukishita, K. Lee, S. Kim, Y. Yumoto, A. Kobayashi, T. Shirasawa, and H. Kobayashi, “Age and Sex-Dependent Alterations in Heart Rate Variability Profiling the Characteristics of Men and Women in Their 30s,” Anti-Aging Medicine, Vol.7, No.8, pp. 94-99, 2010. https://doi.org/10.3793/jaam.7.94
- [4] R. K. Tripathy, A. Acharya, and S. K. Choudhary, “Gender Classification from ECG Signal Analysis Using Least Square Support Vector Machine,” American J. of Signal Processing, Vol.2, No.5, pp. 145-149, 2012. https://doi.org/10.5923/j.ajsp.20120205.08
- [5] J. Hayano, K. Kisohara, Y. Yoshida, H. Sakano, and E. Yuda, “Association of heart rate variability with regional difference in senility death ratio: ALLSTAR big data analysis,” SAGE Open Med., Vol.7, Article No.2050312119852259, 2019. https://doi.org/10.1177/2050312119852259
- [6] J. Hayano, E. Yuda, and Y. Yoshida, “Association of 24-hour heart rate variability and daytime physical activity ALLSTAR big data analysis,” Int. J. of Bioscience, Biochemistry and Bioinformatics, Vol.8, No.1, pp. 61-67, 2018. https://doi.org/10.17706/ijbbb.2018.8.1.61-67
- [7] J. Hayano, N. Ueda, K. Kisohara, E. Yuda, R. M. Carney, and J. A. Blumenthal, “Survival predictors of heart rate variability after myocardial infarction with and without low left ventricular ejection fraction,” Front. Neurosci., Vol.15, Articel No.610955, 2021. https://doi.org/10.3389/fnins.2021.610955
- [8] E. Yuda, N. Ueda, M. Kisohara, and J. Hayano, “Redundancy among risk predictors derived from heart rate variability and dynamics: ALLSTAR big data analysis,” Annals of Nnoninvasive Electrocardiology, Vol.26, Issue 1, Article No.e12790, 2021. https://doi.org/10.1111/anec.12790
- [9] X. Xu and H. Liu, “ECG Heartbeat Classification Using Convolutional Neural Networks,” IEEE Access, Vol.8, pp. 8614-8619, 2020. https://doi.org/10.1109/ACCESS.2020.2964749
- [10] “Allostatic State Mapping by Ambulatory ECG Repository (ALLSTAR).” http://www.med.nagoya-cu.ac.jp/mededu.dir/allstar/ [Accessed April 28, 2022]
- [11] J. Hayano, E. Watanabe, Y. Saito, F. Sasaki, K. Fujimoto, T. Nomiyama, K. Kawai, I. Kodama, and H. Sakakibara, “Screening for Obstructive Sleep Apnea by Cyclic Variation of Heart Rate,” Circulation: Arrhythmia and Electrophysiology, Vol.4, No.1, pp. 64-72, 2011. https://doi.org/10.1161/CIRCEP.110.958009
- [12] O. Faust, R. Barika, A. Shenfield, E. J. Ciaccio, and U. Acharya, “Accurate detection of sleep apnea with long short-term memory network based on RR interval signals,” Knowl.-Based Syst., Vol.212, Article No.106591, 2021. https://doi.org/10.1016/j.knosys.2020.106591
- [13] Q. Shen, H. Qin, K. Wei, and G. Liu, “Multiscale Deep Neural Network for Obstructive Sleep Apnea Detection Using RR Interval from Single-Lead ECG Signal,” IEEE Trans. on Instrumentation and Measurement, Vol.70, Article No.3062414, 2021. https://doi.org/10.1109/TIM.2021.3062414
- [14] L. Wang, Y. Lin, and J. Wang, “A RR interval based automated apnea detection approach using residual network,” Computer Methods and Programs in Biomedicine, Vol.176, pp. 93-104, 2019. https://doi.org/10.1016/j.cmpb.2019.05.002
- [15] C. Dwork and A. Roth, “The Algorithmic Foundations of Differential Privacy,” Found. Trends Theor. Comput. Sci., Vol.9, Issues 3-4, pp. 211-407, 2014. https://doi.org/10.1561/0400000042
- [16] E. Bozkir, O. Günlü, W. Fuhl, R. F. Schaefer, and E. Kasneci, “Differential privacy for eye tracking with temporal correlations,” PLoS ONE, Vol.16, No.8, Article No.e0255979, 2021. https://doi.org/10.1371/journal.pone.0255979
- [17] P. Kairouz, S. Oh, and P. Viswanath, “Extremal Mechanisms for Local Differential Privacy,” arXiv:1407.1338v3, 2014. https://doi.org/10.48550/arXiv.1407.1338
- [18] U. Erlingsson, V. Pihur, and A. Korolova, “RAPPOR: Randomized Aggregatable Privacy-Preserving Ordinal Response,” Proc. of the 2014 ACM SIGSAC Conf. on Computer and Communications Security (CCS ’14), pp. 1054-1067, 2014. https://doi.org/10.1145/2660267.2660348
 This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.
This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.