Paper:
Image-Based Gel Encapsulation of Suspended Single Cells for Parallel Single-Cell Screening
Venkatesh Kumar Panneer Selvam*, Muhammad Luqman Arief Bin Kamaludin*, Ghulam Murtaza*, Rifat Hussain Chowdhury*, Tanmay Debnath*, Shunya Okamoto*, Takayuki Shibata*
 , Tuhin Subhra Santra**
, Tuhin Subhra Santra**
 , and Moeto Nagai*,***
, and Moeto Nagai*,***

*Department of Mechanical Engineering, Toyohashi University of Technology
1-1 Hibarigaoka, Tempaku-cho, Toyohashi, Aichi 441-8580, Japan
**Department of Engineering Design, Indian Institute of Technology
Chennai, Tamil Nadu 600, India
***Institute for Research on Next-generation Semiconductor and Sensing Science, Toyohashi University of Technology
1-1 Hibarigaoka, Tempaku-cho, Toyohashi, Aichi 441-8580, Japan
Single-cell screening, which has revolutionized the life sciences, is an important method for detecting, separating, or treating specific cells based on desired characteristics. Previously, single cells of interest were manually identified in an image, which required human labor and time. We developed an automated photopolymerization system to encapsulate suspended single cells in approximately 50-µm photo-crosslinkable hydrogel squares. An image was captured, and single cells were selected from grouped cells based on image processing. A generated image was transferred to a digital micromirror device (DMD), and in parallel, target-suspended single cells were encapsulated in gelatin methacryloyl (GelMA) hydrogels. We built a data transfer platform based on a Power Automate Desktop (PAD), completed the data transfer, and projected the processed image onto a sample in 10 s, ensuring a minimum alignment error of 6.2 µm.
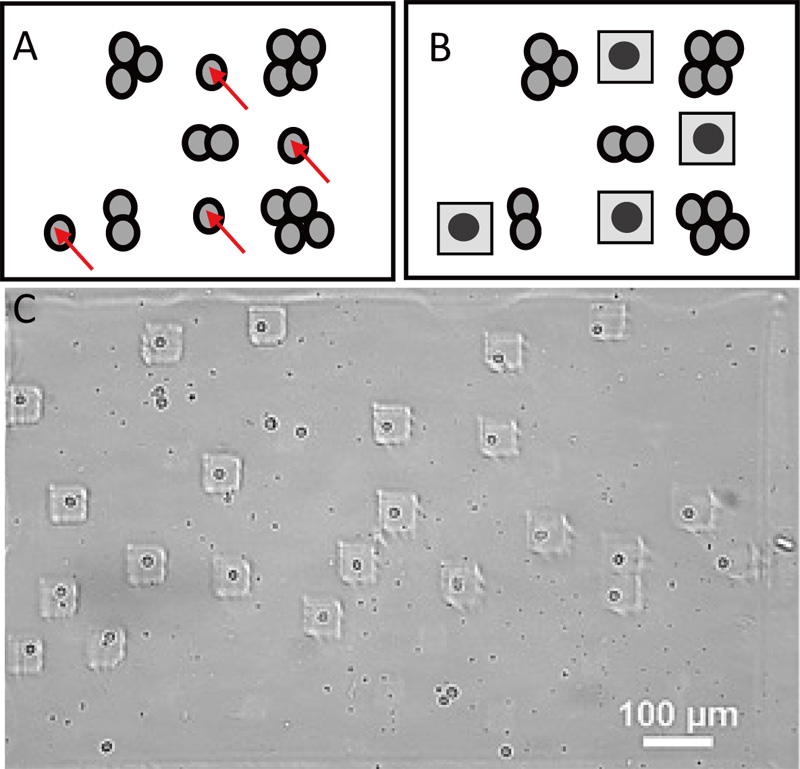
Image-based gel encapsulation of suspended single cells
- [1] L. Valihrach, P. Androvic, and M. Kubista, “Platforms for single-cell collection and analysis,” Int. J. of Molecular Sciences, Vol.19, No.3, Article No.807, 2018. https://doi.org/10.3390/ijms19030807
- [2] A. Dalili, E. Samiei, and M. Hoorfar, “A review of sorting, separation and isolation of cells and microbeads for biomedical applications: Microfluidic approaches,” Analyst, Vol.144, No.1, pp. 87-113, 2019. https://doi.org/10.1039/C8AN01061G
- [3] V. Fitzgerald and P. Leonard, “Single cell screening approaches for antibody discovery,” Methods, Vol.116, pp. 34-42, 2017. https://doi.org/10.1016/j.ymeth.2016.11.006
- [4] N. B. Ben-Moshe et al., “Predicting bacterial infection outcomes using single cell RNA-sequencing analysis of human immune cells,” Nature Communications, Vol.10, No.1, Article No.3266, 2019. https://doi.org/10.1038/s41467-019-11257-y
- [5] C. Kim et al., “Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing,” Cell, Vol.173, No.4, pp. 879-893.e13, 2018. https://doi.org/10.1016/j.cell.2018.03.041
- [6] A. K. Mitra et al., “Single-cell analysis of targeted transcriptome predicts drug sensitivity of single cells within human myeloma tumors,” Leukemia, Vol.30, No.5, pp. 1094-1102, 2016. https://doi.org/10.1038/leu.2015.361
- [7] B. E. Debs, R. Utharala, I. V. Balyasnikova, A. D. Griffiths, and C. A. Merten, “Functional single-cell hybridoma screening using droplet-based microfluidics,” Proc. of the National Academy of Sciences, Vol.109, No.29, pp. 11570-11575, 2012. https://doi.org/10.1073/pnas.1204514109
- [8] C. Krieg et al., “High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy,” Nature Medicine, Vol.24, No.2, pp. 144-153, 2018. https://doi.org/10.1038/nm.4466
- [9] M. B. Dainiak, A. Kumar, I. Y. Galaev, and B. Mattiasson, “Methods in cell separations,” A. Kumar, I. Y. Galaev, and B. Mattiasson (Eds.), “Cell separation: Fundamentals, analytical and preparative methods,” Springer Berlin Heidelberg, pp. 1-18, 2007.
- [10] P. Shinde et al., “Current trends of microfluidic single-cell technologies,” Int. J. of Molecular Sciences, Vol.19, No.10, Article No.3143, 2018. https://doi.org/10.3390/ijms19103143
- [11] X. Liao, M. Makris, and X. M. Luo, “Fluorescence-activated cell sorting for purification of plasmacytoid dendritic cells from the mouse bone marrow,” J. of Visualized Experiments, No.117, Article No.54641, 2016. https://doi.org/10.3791/54641
- [12] A. Blasius, W. Vermi, A. Krug, F. Facchetti, M. Cella, and M. Colonna, “A cell-surface molecule selectively expressed on murine natural interferon-producing cells that blocks secretion of interferon-alpha,” Blood, Vol.103, No.11, pp. 4201-4206, 2004. https://doi.org/10.1182/blood-2003-09-3108
- [13] A. Gross, J. Schoendube, S. Zimmermann, M. Steeb, R. Zengerle, and P. Koltay, “Technologies for single-cell isolation,” Int. J. of Molecular Sciences, Vol.16, No.8, pp. 16897-16919, 2015. https://doi.org/10.3390/ijms160816897
- [14] G. Welzel, D. Seitz, and S. Schuster, “Magnetic-activated cell sorting (MACS) can be used as a large-scale method for establishing zebrafish neuronal cell cultures,” Scientific Reports, Vol.5, No.1, Article No.7959, 2015. https://doi.org/10.1038/srep07959
- [15] H. Valli et al., “Fluorescence- and magnetic-activated cell sorting strategies to isolate and enrich human spermatogonial stem cells,” Fertility and Sterility, Vol.102, No.2, pp. 566-580.e7, 2014. https://doi.org/10.1016/j.fertnstert.2014.04.036
- [16] S. Ota et al., “Ghost cytometry,” Science, Vol.360, No.6394, pp. 1246-1251, 2018. https://doi.org/10.1126/science.aan0096
- [17] N. Nitta et al., “Intelligent image-activated cell sorting,” Cell, Vol.175, No.1, pp. 266-276.e13, 2018. https://doi.org/10.1016/j.cell.2018.08.028
- [18] C. A. LaBelle, A. Massaro, B. Cortés-Llanos, C. E. Sims, and N. L. Allbritton, “Image-based live cell sorting,” Trends in Biotechnology, Vol.39, No.6, pp. 613-623, 2021. https://doi.org/10.1016/j.tibtech.2020.10.006
- [19] B. Cortés-Llanos, Y. Wang, C. E. Sims, and N. L. Allbritton, “A technology of a different sort: Microraft arrays,” Lab on a Chip, Vol.21, No.17, pp. 3204-3218, 2021. https://doi.org/10.1039/D1LC00506E
- [20] J. Jin et al., “Robotic data acquisition with deep learning enables cell image-based prediction of transcriptomic phenotypes,” Proc. of the National Academy of Sciences, Vol.120, No.1, Article No.e2210283120, 2022. https://doi.org/10.1073/pnas.2210283120
- [21] M. Nagai, K. Kato, K. Oohara, and T. Shibata, “Pick-and-place operation of single cell using optical and electrical measurements for robust manipulation,” Micromachines, Vol.8, No.12, Article No.350, 2017. https://doi.org/10.3390/mi8120350
- [22] Y. Yamanishi, T. Nakano, Y. Sawada, K. Itoga, T. Okano, and F. Arai, “Maskless gray scale lithography and its 3d microfluidic applications,” J. Robot. Mechatron., Vol.23, No.3, pp. 426-433, 2011. https://doi.org/10.20965/jrm.2011.p0426
- [23] M. Nagai, K. Oohara, K. Kato, T. Kawashima, and T. Shibata, “Development and characterization of hollow microprobe array as a potential tool for versatile and massively parallel manipulation of single cells,” Biomedical Microdevices, Vol.17, No.2, Article No.41, 2015. https://doi.org/10.1007/s10544-015-9943-z
- [24] M. Nagai, K. Kato, S. Soga, T. S. Santra, and T. Shibata, “Scalable parallel manipulation of single cells using micronozzle array integrated with bidirectional electrokinetic pumps,” Micromachines, Vol.11, No.4, Article No.442, 2020. https://doi.org/10.3390/mi11040442
- [25] R. Negishi et al., “Gel-based cell manipulation method for isolation and genotyping of single-adherent cells,” Analyst, Vol.144, No.3, pp. 990-996, 2019. https://doi.org/10.1039/C8AN01456F
- [26] T. Yoshino, T. Tanaka, S. Nakamura, R. Negishi, M. Hosokawa, and T. Matsunaga, “Manipulation of a single circulating tumor cell using visualization of hydrogel encapsulation toward single-cell whole-genome amplification,” Analytical Chemistry, Vol.88, No.14, pp. 7230-7237, 2016. https://doi.org/10.1021/acs.analchem.6b01475
- [27] R. Negishi, K. Takai, T. Tanaka, T. Matsunaga, and T. Yoshino, “High-throughput manipulation of circulating tumor cells using a multiple single-cell encapsulation system with a digital micromirror device,” Analytical Chemistry, Vol.90, No.16, pp. 9734-9741, 2018. https://doi.org/10.1021/acs.analchem.8b00896
- [28] M. B. Browning, S. N. Cereceres, P. T. Luong, and E. M. Cosgriff-Hernandez, “Determination of the in vivo degradation mechanism of PEGDA hydrogels,” J. of Biomedical Materials Research Part A, Vol.102, No.12, pp. 4244-4251, 2014. https://doi.org/10.1002/jbm.a.35096
- [29] M. Zhu, Y. Wang, G. Ferracci, J. Zheng, N.-J. Cho, and B. H. Lee, “Gelatin methacryloyl and its hydrogels with an exceptional degree of controllability and batch-to-batch consistency,” Scientific Reports, Vol.9, No.1, Article No.6863, 2019. https://doi.org/10.1038/s41598-019-42186-x
- [30] M. Nagai et al., “Parallel photothermal coalescence of biocompatible photocurable PEGDA droplets,” IEEJ Trans. on Sensors and Micromachines, Vol.143, No.3, pp. 49-54, 2023. https://doi.org/10.1541/ieejsmas.143.49
- [31] T. Debnath, R. Hattori, S. Okamoto, T. Shibata, T. S. Santra, and M. Nagai, “Automated detection of patterned single-cells within hydrogel using deep learning,” Scientific Reports, Vol.12, No.1, Article No.18343, 2022. https://doi.org/10.1038/s41598-022-22774-0
 This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.
This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.