Review:
Burnt-in Text Recognition from Medical Imaging Modalities: Existing Machine Learning Practices
Efosa Osagie†, Wei Ji, and Na Helian

Department of Computer Science, University of Hertfordshire
College Lane Campus, Hatfield, Hertfordshire AL 9, United Kingdom
†Corresponding author
In recent times, medical imaging has become a significant component of clinical diagnosis and examinations to detect and evaluate various medical conditions. The interpretation of these medical examinations and the patient’s demographics are usually textual data, which is burned in on the pixel content of medical imaging modalities (MIM). Example of these MIM includes ultrasound and X-ray imaging. As artificial intelligence advances for medical applications, there is a high demand for the accessibility of these burned-in textual data for various needs. This article aims to review the significance of burned-in textual data recognition in MIM and recent research regarding the machine learning approach, challenges, and open issues for further investigation on this application. The review describes the significant problems in this study area as low resolution and background interference of textual data. Finally, the review suggests applying more advanced deep learning ensemble algorithms as possible solutions.
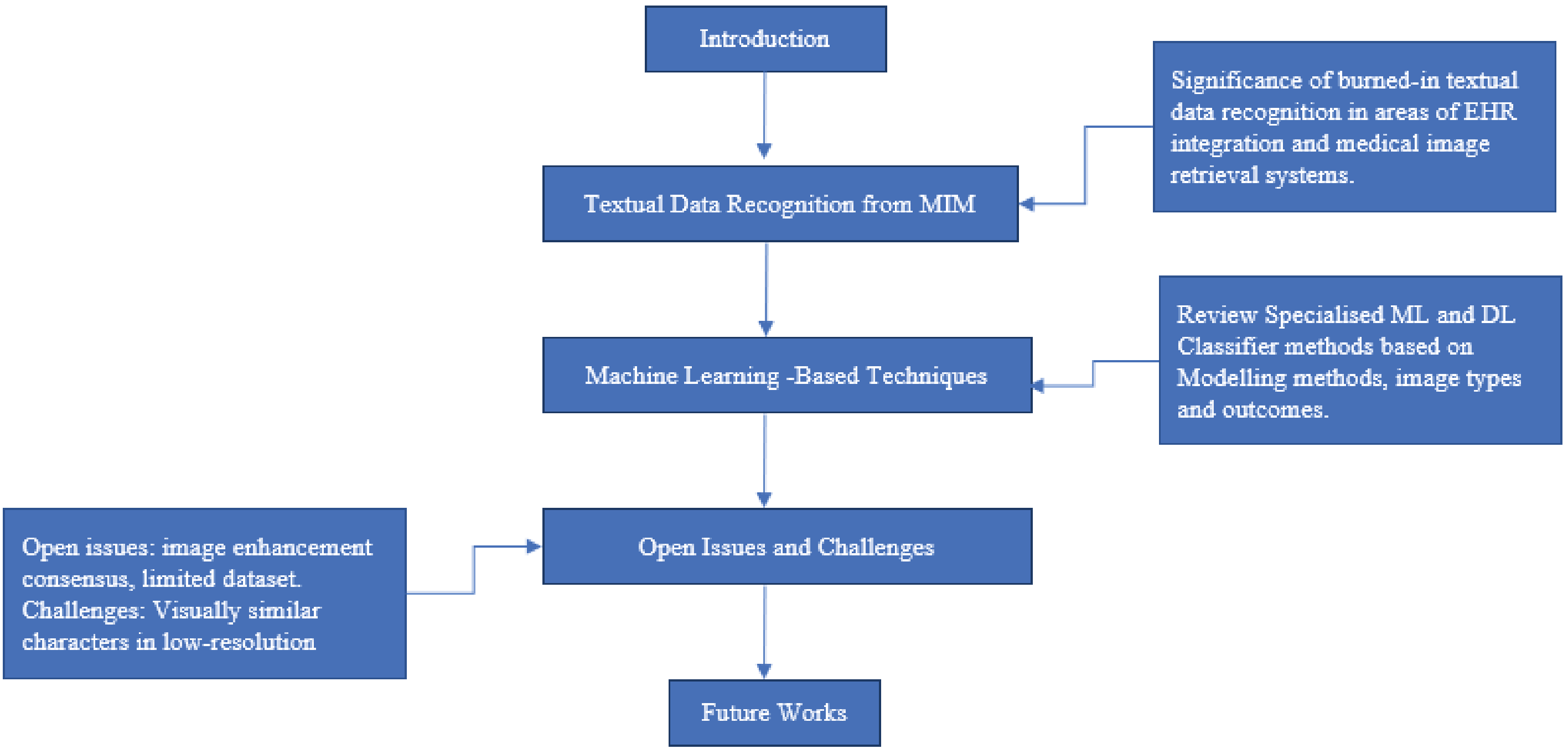
Medical image character recognition using deep learning
- [1] K. Baskar, “A survey on feature selection techniques in medical image processing,” Computer Science, 2018. https://www.semanticscholar.org/paper/A-Survey-on-Feature-Selection-Techniques-in-Medical-Baskar/280694439253fc179a5a4157af18f09177af105c [Accessed November 6, 2022]
- [2] J. Z. Wang, “Security filtering of medical images using OCR,” Proc. of the 3rd All-Russian Scientific Conf., pp. 118-122, 2001.
- [3] T. Davenport and R. Kalakota, “The potential for artificial intelligence in healthcare,” Future Healthcare J., Vol.6, No.2, pp. 94-98, 2019. https://doi.org/10.7861/futurehosp.6-2-94
- [4] C. B. Collin et al., “Computational models for clinical applications in personalized medicine—guidelines and recommendations for data integration and model validation,” J. of Personalized Medicine, Vol.12, No.2, Article No.166, 2022. https://doi.org/10.3390/jpm12020166
- [5] V. Ehrenstein et al., “Obtaining data from electronic health records,” R. E. Gliklich, M. B. Leavy, and N. A. Dreyer (Eds.), “Tools and Technologies for Registry Interoperability, Registries for Evaluating Patient Outcomes: A User’s Guide, 3rd Edition, Addendum 2,” Agency for Healthcare Research and Quality, 2019. https://www.ncbi.nlm.nih.gov/books/NBK551878/ [Accessed November 12, 2022]
- [6] A. Paulsen, S. Overgaard, and J. M. Lauritsen, “Quality of data entry using single entry, double entry and automated forms processing—an example based on a study of patient-reported outcomes,” PLOS ONE, Vol.7, No.4, Article No.e35087, 2012. https://doi.org/10.1371/journal.pone.0035087
- [7] R. J. McDonald et al., “The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload,” Academic Radiology, Vol.22, No.9, pp. 1191-1198, 2015. https://doi.org/10.1016/j.acra.2015.05.007
- [8] B. Bergeron, “Clinical data capture: OMR and OCR and your flatbed scanner,” MedGenMed, Vol.7, No.2, Article No.66, 2005.
- [9] R. Rabiei et al., “Prediction of breast cancer using machine learning approaches,” J. of Biomedical Physics and Engineering, Vol.12, No.3, pp. 297-308, 2022. https://doi.org/10.31661/jbpe.v0i0.2109-1403
- [10] Y. Amethiya et al., “Comparative analysis of breast cancer detection using machine learning and biosensors,” Intelligent Medicine, Vol.2, No.2, pp. 69-81, 2022. https://doi.org/10.1016/j.imed.2021.08.004
- [11] W. Newhauser et al., “Anonymization of DICOM electronic medical records for radiation therapy,” Computers in Biology and Medicine, Vol.53, pp. 134-140, 2014. https://doi.org/10.1016/j.compbiomed.2014.07.010
- [12] E. Menasalvas and C. Gonzalo-Martin, “Challenges of medical text and image processing: Machine learning approaches,” A. Holzinger (Ed.), “Machine Learning for Health Informatics: State-of-the-Art and Future Challenges,” pp. 221-242, Springer, 2016. https://doi.org/10.1007/978-3-319-50478-0_11
- [13] H. Kawano et al., “Structure extraction from decorated characters by graph spectral decomposition and component selection criterion,” J. Adv. Comput. Intell. Intell. Inform., Vol.14, No.2, pp. 179-184, 2010. https://doi.org/10.20965/jaciii.2010.p0179
- [14] H. Miyao et al., “Printed Japanese character recognition using multiple commercial OCRs,” J. Adv. Comput. Intell. Intell. Inform., Vol.8, No.2, pp. 200-207, 2004. https://doi.org/10.20965/jaciii.2004.p0200
- [15] J. Park et al., “Multi-lingual optical character recognition system using the reinforcement learning of character segmenter,” IEEE Access, Vol.8, pp. 174437-174448, 2020. https://doi.org/10.1109/ACCESS.2020.3025769
- [16] P. Vcelak et al., “Identification and classification of DICOM files with burned-in text content,” Int. J. of Medical Informatics, Vol.126, pp. 128-137, 2019. https://doi.org/10.1016/j.ijmedinf.2019.02.011
- [17] K. Mohsenzadegan, V. Tavakkoli, and K. Kyamakya, “Deep neural network concept for a blind enhancement of document-images in the presence of multiple distortions,” Applied Sciences, Vol.12, No.19, Article No.9601, 2022. https://doi.org/10.3390/app12199601
- [18] G. K. Tsui and T. Chan, “Automatic selective removal of embedded patient information from image content of DICOM files,” American J. of Roentgenology, Vol.198, No.4, pp. 769-772, 2012. https://doi.org/10.2214/AJR.10.6352
- [19] E. Monteiro, C. Costa, and J. L. Oliveira, “A machine learning methodology for medical imaging anonymization,” 2015 37th Annual Int. Conf. of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 1381-1384, 2015. https://doi.org/10.1109/EMBC.2015.7318626
- [20] Y. Ma and Y. Wang, “Text detection in medical images using local feature extraction and supervised learning,” 2015 12th Int. Conf. on Fuzzy Systems and Knowledge Discovery (FSKD), pp. 953-958, 2015. https://doi.org/10.1109/FSKD.2015.7382072
- [21] C. Reul et al., “Expectation-driven text extraction from medical ultrasound images,” A. Hoerbst et al. (Eds.), “Exploring Complexity in Health: An Interdisciplinary Systems Approach,” pp. 712-716, IOS Press, 2016. https://doi.org/10.3233/978-1-61499-678-1-712
- [22] E. Monteiro, C. Costa, and J. L. Oliveira, “A de-identification pipeline for ultrasound medical images in DICOM format,” J. of Medical Systems, Vol.41, No.5, Article No.89, 2017. https://doi.org/10.1007/s10916-017-0736-1
- [23] J. M. Silva et al., “Controlled searching in reversibly de-identified medical imaging archives,” J. of Biomedical Informatics, Vol.77, pp. 81-90, 2018. https://doi.org/10.1016/j.jbi.2017.12.002
- [24] X. Xu, W. Wang, and Q. Liu, “Medical image character recognition based on multi-scale neural convolutional network,” 2021 Int. Conf. on Security, Pattern Analysis, and Cybernetics (SPAC), pp. 408-412, 2021. https://doi.org/10.1109/SPAC53836.2021.9539999
- [25] M. Antunes, R. Machado, and A. Silva, “Anonymization of burned-in annotations in ultrasound imaging,” Eletrónica e Telecomunicações, Vol.5, No.3, pp. 360-364, 2011.
- [26] J. P. Segal and R. Hansen, “Medical images, social media and consent,” Nature Reviews Gastroenterology & Hepatology, Vol.18, No.8, pp. 517-518, 2021. https://doi.org/10.1038/s41575-021-00453-1
- [27] A. Badano et al., “Consistency and standardization of color in medical imaging: A consensus report,” J. of Digital Imaging, Vol.28, No.1, pp. 41-52, 2015. https://doi.org/10.1007/s10278-014-9721-0
- [28] M. Kociołek, M. Strzelecki, and R. Obuchowicz, “Does image normalization and intensity resolution impact texture classification?,” Computerized Medical Imaging and Graphics, Vol.81, Article No.101716, 2020. https://doi.org/10.1016/j.compmedimag.2020.101716
- [29] L. Maier-Hein et al., “Why rankings of biomedical image analysis competitions should be interpreted with care,” Nature Communications, Vol.9, No.1, Article No.5217, 2018. https://doi.org/10.1038/s41467-018-07619-7
- [30] M. Aljabri et al., “Towards a better understanding of annotation tools for medical imaging: A survey,” Multimedia Tools and Applications, Vol.81, No.18, pp. 25877-25911, 2022. https://doi.org/10.1007/s11042-022-12100-1
- [31] Y. Li, B. Sixou, and F. Peyrin, “A review of the deep learning methods for medical images super resolution problems,” IRBM, Vol.42, No.2, pp. 120-133, 2021. https://doi.org/10.1016/j.irbm.2020.08.004
- [32] H. Michalak and K. Okarma, “Improvement of image binarization methods using image preprocessing with local entropy filtering for alphanumerical character recognition purposes,” Entropy, Vol.21, No.6, Article No.562, 2019. https://doi.org/10.3390/e21060562
- [33] W. Bieniecki, S. Grabowski, and W. Rozenberg, “Image preprocessing for improving OCR accuracy,” 2007 Int. Conf. on Perspective Technologies and Methods in MEMS Design, pp. 75-80, 2007. https://doi.org/10.1109/MEMSTECH.2007.4283429
- [34] S. Nomura et al., “Morphological preprocessing method to thresholding degraded word images,” Pattern Recognition Letters, Vol.30, No.8, pp. 729-744, 2009. https://doi.org/10.1016/j.patrec.2009.03.008
- [35] J. M. D. Delgado and L. Oyedele, “Deep learning with small datasets: Using autoencoders to address limited datasets in construction management,” Applied Soft Computing, Vol.112, Article No.107836, 2021. https://doi.org/10.1016/j.asoc.2021.107836
- [36] M. Li, R. Poovendran, and S. Narayanan, “Protecting patient privacy against unauthorized release of medical images in a group communication environment,” Computerized Medical Imaging and Graphics, Vol.29, No.5, pp. 367-383, 2005. https://doi.org/10.1016/j.compmedimag.2005.02.003
- [37] X. Qin, F. M. Bui, and H. H. Nguyen, “Learning from an imbalanced and limited dataset and an application to medical imaging,” 2019 IEEE Pacific Rim Conf. on Communications, Computers and Signal Processing (PACRIM), 2019. https://doi.org/10.1109/PACRIM47961.2019.8985057
- [38] D. Pal et al., “MSHSCNN: Multi-scale hybrid-Siamese network to differentiate visually similar character classes,” 2021 9th European Workshop on Visual Information Processing (EUVIP), 2021. https://doi.org/10.1109/EUVIP50544.2021.9483980
- [39] P. Inkeaw et al., “Recognition of similar characters using gradient features of discriminative regions,” Expert Systems with Applications, Vol.134, pp. 120-137, 2019. https://doi.org/10.1016/j.eswa.2019.05.050
- [40] A. Safaei, “Text-based multi-dimensional medical images retrieval according to the features-usage correlation,” Medical & Biological Engineering & Computing, Vol.59, No.10, pp. 1993-2017, 2021. https://doi.org/10.1007/s11517-021-02392-0
- [41] D. Cireşan, U. Meier, and J. Schmidhuber, “Multi-column deep neural networks for image classification,” arXiv: 1202.2745, 2012. https://doi.org/10.48550/ARXIV.1202.2745
- [42] A. Krizhevsky, I. Sutskever, and G. E. Hinton, “ImageNet classification with deep convolutional neural networks,” Communications of the ACM, Vol.60, No.6, pp. 84-90, 2017. https://doi.org/10.1145/3065386
- [43] L. Lam and S. Y. Suen, “Application of majority voting to pattern recognition: An analysis of its behavior and performance,” IEEE Trans. on Systems, Man, and Cybernetics – Part A: Systems and Humans, Vol.27, No.5, pp. 553-568, 1997. https://doi.org/10.1109/3468.618255
- [44] J. Shlens, “A tutorial on principal component analysis,” arXiv: 1404.1100, 2014. https://doi.org/10.48550/ARXIV.1404.1100
- [45] Z. M. Kovács-V, “A novel architecture for high quality hand-printed character recognition,” Pattern Recognition, Vol.28, No.11, pp. 1685-1692, 1995. https://doi.org/10.1016/0031-3203(95)00044-Z
 This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.
This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.