Paper:
A Kernel k-Means-Based Method and Attribute Selections for Diabetes Diagnosis
Tru Cao*,**, Chau Vo*, Son Nguyen*, Atsushi Inoue**, and Duanning Zhou**
*Ho Chi Minh City University of Technology, Vietnam National University
268 Ly Thuong Kiet Street, District 10, Ho Chi Minh City, Vietnam
**Eastern Washington University
668 North Riverpoint Boulevard, Spokane, Washington 99202-1677, USA
Diabetes diagnosis is important due to the high death rate and complication consequences caused by the disease. First, we propose a kernel k-means-based prediction method and explore attribute selections for effective and robust diabetes diagnosis. This method derives homogeneous sub-clusters in the high dimensional kernelized feature space to compute the distance of a new instance to those sub-clusters, and then apply the 1-nearest neighbor to classify it as positive or negative to the disease. Our experimental results could identify the best effective attribute group for each considered prediction method and show that the proposed method outperforms the existing ones for the task. Second, we introduce our developed diabetes visualization and decision support system, named DIAVIS, which is equipped with the proposed prediction method. This system can support doctors to diagnose diabetes and track patient health progress to prescribe proper medications in a treatment process.
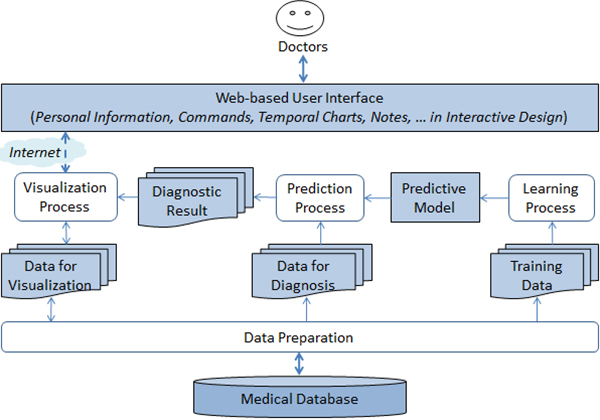
Overview architecture of DIAVIS system
- [1] World Health Organization (WHO), “Diabetes,” http://www.who.int/mediacentre/factsheets/fs312/en/ [accessed February 19, 2018]
- [2] M. Pohl, S. Wiltner, A. Rind, W. Aigner, S. Miksch, T. Turic, and F. Drexler, “Patient development at a glance: an evaluation of a medical data visualization,” IFIP Conf. on Human-Computer Interaction, LNCS Vol.6949, pp. 292-299, 2011.
- [3] D. Bertsimas, N. Kallus, A. M. Weinstein, and Y. D. Zhuo, “Personalized diabetes management using electronic medical records,” Diabetes Care, Vol.40, No.2, pp. 210-217, 2017.
- [4] D. K. Choubey and S. Paul, “Classification techniques for diagnosis of diabetes: a review,” Int. J. Biomedical Engineering and Technology, Vol.21, No.1, pp. 15-39, 2016.
- [5] Y. Hayshi and S. Yukita, “Rule extraction using recursive-rule extraction algorithm with J48graft combined with sampling selection techniques for the diagnosis of type 2 diabetes mellitus in the PIMA Indian dataset,” Informatics in Medicine Unlocked, Vol.2, pp. 92-104, 2016.
- [6] K. V. S. R. P. Varma, A. A. Rao, T. S. Mahalakshmi, and P. V. N. Rao, “A computational intelligence technique for the effective diagnosis of diabetic patients using Principal Component Analysis (PCA) and modified fuzzy SLIQ decision tree approach,” Applied Soft Computing, Vol.49, pp. 137-145, 2016.
- [7] S. Bahramian and A. Nikravanshalmani, “Hybrid algorithm based on k-nearest-neighbor algorithm and Adaboost with selection of feature by genetic algorithm for the diagnosis of diabetes,” Int. J. of Mechatronics, Electrical and Computer Technology, Vol.6, No.21, pp. 2977-2986, 2016.
- [8] N. Nai-arun and R. Moungmai, “Comparison of classifiers for the risk of diabetes prediction,” Procedia Computer Science, Vol.69, pp. 132-142, 2015.
- [9] S. Ramesh, H. Balaji, N. Ch. S. N. Iyengar, and R. D. Caytiles, “Optimal predictive analytics of Pima diabetics using deep learning,” Int. J. of Database Theory and Application, Vol.10, No.9, pp. 47-62, 2017.
- [10] D. K. Choubey and S. Paul, “GA_RBF NN: a classification system for diabetes,” Int. J. of Biomedical Engineering and Technology, Vol.23, No.1, pp. 71-93, 2017.
- [11] O. Erkaymaz, M. Ozer, and M. Perc, “Performance of small-world feedforward neural networks for the diagnosis of diabetes,” Applied Mathematics and Computation, Vol.311, pp. 22-28, 2017.
- [12] N. Khan, D. Gaurav, and T. Kandl, “Performance evaluation of Levenberg-Marquardt technique in error reduction for diabetes condition classification,” Procedia Computer Science, Vol.18, pp. 2629-2637, 2013.
- [13] M. Nilashi, O. Ibrahim, M. Dalvi, H. Ahmadi, and L. Shahmoradi, “Accuracy improvement for diabetes disease classification: a case on a public medical dataset,” Fuzzy Inf. Eng., Vol.9, No.3, pp. 345-357, 2017.
- [14] J. W. Smith, J. E. Everhart, W. C. Dickson, W. C. Knowler, and R. S. Johannes, “Using the ADAP learning algorithm to forecast the onset of diabetes mellitus,” Proc Annu Symp Comput Appl Med Care, pp. 261-265, 1988.
- [15] A. H. Osman and H. M. Aljahdali, “Diabetes disease diagnosis method based on feature extraction using K-SVM,” Int. J. of Advanced Computer Science and Applications, Vol.8, No.1, pp. 236-244, 2017.
- [16] T. Santhanam and M. S. Padmavathi, “Application of k-means and genetic algorithms for dimension reduction by integrating SVM for diabetes diagnosis,” Procedia Computer Science, Vol.47, pp. 76-83, 2015.
- [17] S. Lekkas and L. Mikhailov, “Evolving fuzzy medical diagnosis of Pima Indians diabetes and of dermatological diseases,” Artificial Intelligence in Medicine, Vol.50, No.2, pp. 117-126, 2010.
- [18] N. Settouti, M. A. Chikh, and M. Saidi, “Generating fuzzy rules for constructing interpretable classifier of diabetes disease,” Australas. Phys. Eng. Sci. Med., Vol.35, No.3, pp. 257-270, 2012.
- [19] MIMIC, “MIMIC-III Critical Care Database,” https://mimic.physionet.org/about/mimic/ [accessed February 26, 2018]
- [20] T. Cao, C. Vo, S. Nguyen, A. Inoue, and D. Zhou, “A kernel k-means-based method for diabetes diagnosis,” Proc. of The ISASE-MAICS 2018, pp. 1-5, 2018.
- [21] G. Tzortzis and A. Likas, “The global kernel k-means clustering algorithm,” 2008 Int. Joint Conf. on Neural Networks, pp. 1977-1984, 2008.
 This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.
This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.