Research Paper:
Minimalist Machine Learning: Binary Classification of Medical Datasets with Matrix Transformations
José Luis Solorio-Ramírez*
 , Oscar Camacho-Nieto**
, Oscar Camacho-Nieto**
 , and Cornelio Yáñez-Márquez*,†
, and Cornelio Yáñez-Márquez*,†

*Centro de Investigacion en Computacion, Instituto Politecnico Nacional
Av. Juan de Dios Batiz S/N, Nueva Industrial Vallejo, Gustavo A. Madero, Ciudad de Mexico 07738, Mexico
†Corresponding author
**Centro de Innovacion y Desarrollo Tecnologico en Computo, Instituto Politecnico Nacional
Av. Juan de Dios Batiz S/N, Nueva Industrial Vallejo, Gustavo A. Madero, Ciudad de Mexico 07738, Mexico
This work introduces an innovative machine learning algorithm based on the minimalist machine learning paradigm, called matrix transformations bootstrap. Evaluated on 15 medical datasets, ranging from 3 to 1626 attributes, the methodology incorporates matrix transformations like rotation and shearing to improve dataset separation in binary classification tasks. Additionally, random feature selection is applied via the bootstrap method, resulting in two new attributes that can be visualized on the Cartesian plane while achieving substantial dimensionality reduction. The results show significant classification performance improvements over traditional algorithms like k-NN, SVM, Bayesian models, ensembles, neural networks, and logistic functions, evaluated using balanced accuracy, recall, and F1-score.
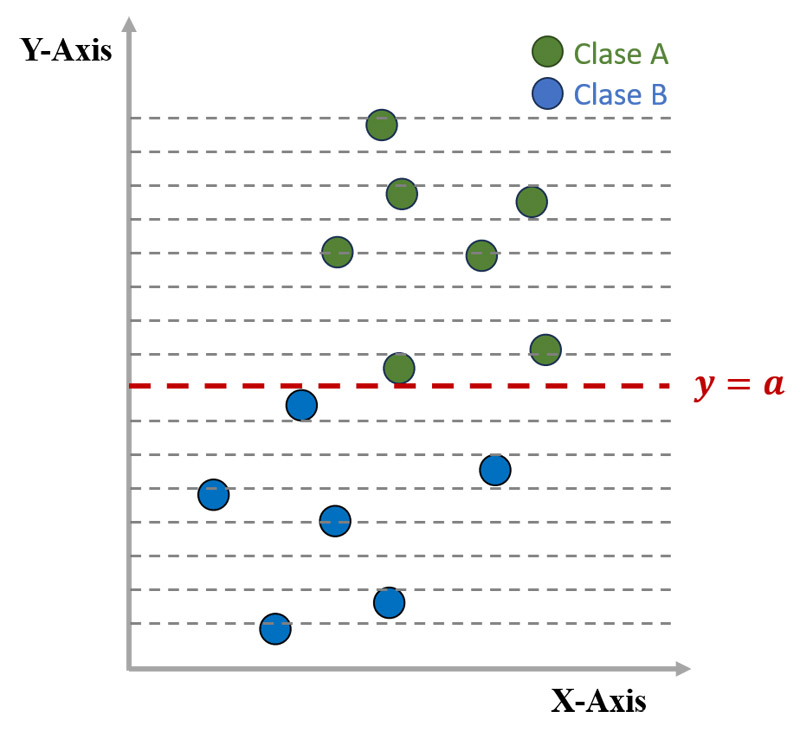
Shift of the decision boundary
- [1] K. Y. Ngiam and I. W. Khor, “Big data and machine learning algorithms for health-care delivery,” The Lancet Oncology, Vol.20, No.5, pp. e262-e273, 2019. https://doi.org/10.1016/S1470-2045(19)30149-4
- [2] C. Iwendi, C. G. Y. Huescas, C. Chakraborty, and S. Mohan, “COVID-19 health analysis and prediction using machine learning algorithms for Mexico and Brazil patients,” J. of Experimental & Theoretical Artificial Intelligence, Vol.36, No.3, pp. 315-335, 2024. https://doi.org/10.1080/0952813X.2022.2058097
- [3] S. Yakubo et al., “Pattern classification in Kampo medicine,” Evidence-Based Complementary and Alternative Medicine, Vol.2014, Article No.535146, 2014. https://doi.org/10.1155/2014/535146
- [4] V. Mhasawade, Y. Zhao, and R. Chunara, “Machine learning and algorithmic fairness in public and population health,” Nature Machine Intelligence, Vol.3, No.8, pp. 659-666, 2021. https://doi.org/10.1038/s42256-021-00373-4
- [5] I. Kononenko, “Machine learning for medical diagnosis: History, state of the art and perspective,” Artificial Intelligence in Medicine, Vol.23, No.1, pp. 89-109, 2001. https://doi.org/10.1016/S0933-3657(01)00077-X
- [6] C. Yáñez-Márquez, “Toward the bleaching of the black boxes: Minimalist machine learning,” IT Professional, Vol.22, No.4, pp. 51-56, 2020. https://doi.org/10.1109/MITP.2020.2994188
- [7] B.-L. Zhang, “Cancer classification by kernel principal component self-regression,” Proc. of the 19th Australian Joint Conf. on Artificial Intelligence, pp. 719-728, 2006. https://doi.org/10.1007/11941439_76
- [8] A. Cüvitoğlu and Z. Işik, “Evaluation machine-learning approaches for classification of cryotherapy and immunotherapy datasets,” Int. J. Machine Learning and Computing, Vol.8, No.4, pp. 331-335, 2018. https://doi.org/10.18178/ijmlc.2018.8.4.707
- [9] M. M. Mijwil and K. Aggarwal, “A diagnostic testing for people with appendicitis using machine learning techniques,” Multimedia Tools and Applications, Vol.81, No.5, pp. 7011-7023, 2022. https://doi.org/10.1007/s11042-022-11939-8
- [10] S. Sharanyaa, P. N. Renjith, and K. Ramesh, “Classification of Parkinson’s disease using speech attributes with parametric and nonparametric machine learning techniques,” 2020 3rd Int. Conf. on Intelligent Sustainable Systems, pp. 437-442, 2020. https://doi.org/10.1109/ICISS49785.2020.9316078
- [11] L. Bergman and Y. Hoshen, “Classification-based anomaly detection for general data,” arXiv:2005.02359, 2020. https://doi.org/10.48550/arXiv.2005.02359
- [12] M. Juez-Gil, Á. Arnaiz-González, J. J. Rodríguez, C. López-Nozal, and C. García-Osorio, “Rotation forest for big data,” Information Fusion, Vol.74, pp. 39-49, 2021. https://doi.org/10.1016/j.inffus.2021.03.007
- [13] J.-L. Solorio-Ramírez, M. Saldana-Perez, M. D. Lytras, M.-A. Moreno-Ibarra, and C. Yáñez-Márquez, “Brain hemorrhage classification in CT scan images using minimalist machine learning,” Diagnostics, Vol.11, No.8, Article No.1449, 2021. https://doi.org/10.3390/diagnostics11081449
- [14] G. Bebis, M. Georgiopoulos, N. da Vitoria Lobo, and M. Shah, “Learning affine transformations,” Pattern Recognition, Vol.32, No.10, pp. 1783-1799, 1999. https://doi.org/10.1016/S0031-3203(98)00178-2
- [15] N. Diaz-Diaz, J. S. Aguilar-Ruiz, J. A. Nepomuceno, and J. Garcia, “Feature selection based on bootstrapping,” 2005 ICSC Congress on Computational Intelligence Methods and Applications, 2005. https://doi.org/10.1109/CIMA.2005.1662338
- [16] F. A. Atienza, J. L. R. Álvarez, G. Camps i Valls, A. Rosado Muñoz, and A. García Alberola, “Bootstrap feature selection in support vector machines for ventricular fibrillation detection,” Proc. of 14th European Symp. on Artificial Neural Networks, pp. 233-238, 2006.
- [17] C. Yüceer and K. Oflazer, “A rotation, scaling, and translation invariant pattern classification system,” Pattern Recognition, Vol.26, No.5, pp. 687-710, 1993. https://doi.org/10.1016/0031-3203(93)90122-D
- [18] S.-W. Chen, X.-S. Wang, and M. Sato, “Uniform polarimetric matrix rotation theory and its applications,” IEEE Trans. on Geoscience and Remote Sensing, Vol.52, No.8, pp. 4756-4770, 2014. https://doi.org/10.1109/TGRS.2013.2284359
- [19] T. Mu, A. K. Nandi, and R. M. Rangayyan, “Classification of breast masses via nonlinear transformation of features based on a kernel matrix,” Medical & Biological Engineering & Computing, Vol.45, No.8, pp. 769-780, 2007. https://doi.org/10.1007/s11517-007-0211-0
- [20] M. B. Richman, “ Rotation of principal components,” J. of Climatology, Vol.6, No.3, pp. 293-335, 1986. https://doi.org/10.1002/joc.3370060305
- [21] R. N. Goldman, “More matrices and transformations: Shear and pseudo-perspective,” J. Arvo (Ed.), “Graphics Gems II,” Morgan Kaufmann, pp. 338-341, 1991. https://doi.org/10.1016/B978-0-08-050754-5.50072-4
- [22] B. Haznedar, M. T. Arslan, and A. Kalinli, “Microarray gene expression cancer data (Version 4),” Mendeley Data, 2017. https://doi.org/10.17632/ynp2tst2hh.4
- [23] “Cervical cancer behavior risk,” UCI Machine Learning Repository, 2019. https://doi.org/10.24432/C5402W
- [24] K. Nakai, “Ecoli,” UCI Machine Learning Repository, 1996. https://doi.org/10.24432/C5388M
- [25] Ş. Gül and F. Rahim, “Period changer,” UCI Machine Learning Repository, 2022. https://doi.org/10.24432/C5B31D
- [26] M. Wang, Y.-Y. Zhang, and F. Min, “Active learning through multi-standard optimization,” IEEE Access, Vol.7, pp. 56772-56784, 2019. https://doi.org/10.1109/ACCESS.2019.2914263
- [27] F. Khozeimeh, R. Alizadehsani, M. Roshanzamir, and P. Layegh, “Immunotherapy dataset,” UCI Machine Learning Repository, 2018. https://doi.org/10.24432/C5DC72
- [28] “Hepatitis,” UCI Machine Learning Repository, 1988. https://doi.org/10.24432/C5Q59J
- [29] S. Haberman, “Haberman’s survival,” UCI Machine Learning Repository, 1999. https://doi.org/10.24432/C5XK51
- [30] F. Khozeimeh, R. Alizadehsani, M. Roshanzamir, P. Layegh, “Cryotherapy dataset,” UCI Machine Learning Repository, 2018. https://doi.org/10.24432/C5FC7C
- [31] M. Amin and A. Ali, “Caesarian section classification dataset,” UCI Machine Learning Repository, 2018. https://doi.org/10.24432/C5N59X
- [32] M. Little, “Parkinsons,” UCI Machine Learning Repository, 2008. https://doi.org/10.24432/C59C74
- [33] K. Nakai, “Yeast,” UCI Machine Learning Repository, 1996. https://doi.org/10.24432/C5KG68
 This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.
This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.