Letter:
Controllable Biological Rhythms and Patterns
Hiroshi Ito*, Takuma Sugi**, and Ken H. Nagai***
*Faculty of Design, Kyushu University
4-9-1 Shiobaru, Fukuoka 815-8540, Japan
**Graduate School of Integrated Sciences for Life, Hiroshima University
1-4-4 Kagamiyama, Higashi-hiroshima, Hiroshima 739-8528, Japan
***School of Materials Science, Japan Advanced Institute of Science and Technology
1-1 Asahidai, Nomi, Ishikawa 923-1292, Japan
One of the goals of soft robotics is to implement intelligent functions capable of processing complex information in soft materials. This is a noble goal, and we already have a familiar example, albeit not an artificial one, in a living organism. We believe that the intelligent biological elements acquired through the evolutionary process, which do not require an electricity supply or CPU, can be used for soft robotics. In this letter, we introduce three biological elements: proteins, squid, and nematodes, which show temporal or special patterns. We then discuss an attempt to apply them to soft robotics.
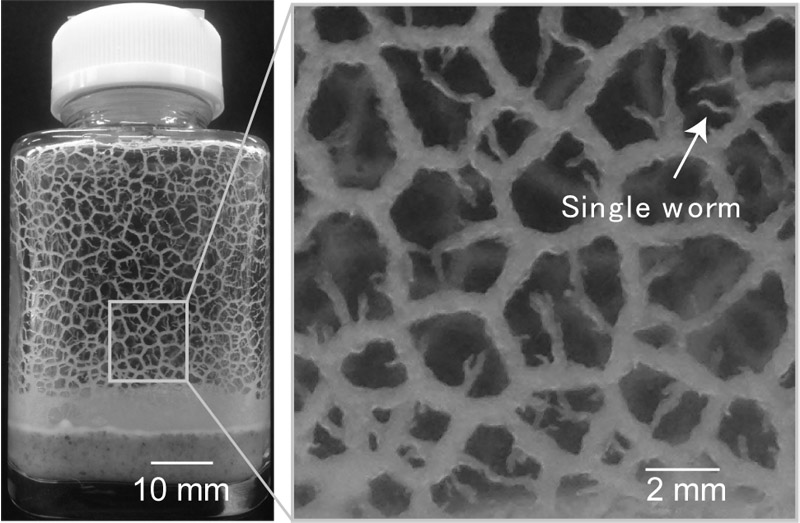
C. elegans population forming a dynamical network
- [1] M. Nakajima, K. Imai, H. Ito, T. Nishiwaki, Y. Murayama, H. Iwasaki, T. Oyama, and T. Kondo, “Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro,” Science, Vol.308, No.5720, pp. 414-415, 2005.
- [2] Y. Murayama, H. Kori, C. Oshima, T. Kondo, H. Iwasaki, and H. Ito, “Low temperature nullifies the circadian clock in cyanobacteria through Hopf bifurcation,” Proc. of National Academy of Sciences, Vol.114, No.22, pp. 5641-5646, 2017.
- [3] T. Yoshida, Y. Murayama, H. Ito, H. Kageyama, and T. Kondo, “Non-parametric entrainment of the in vitro circadian phosphorylation rhythm of cyanobacterial KaiC by temperature cycle,” Proc. of National Academy of Sciences, Vol.106, No.5, pp. 1648-1653, 2009.
- [4] H. Ito, H. Kageyama, M. Mutsuda, M. Nakajima, T. Oyama, and T. Kondo, “Autonomous synchronization of the circadian KaiC phosphorylation rhythm,” Nature Structural and Molecular Biology, Vol.14, pp. 1084-1088, 2007.
- [5] T. Sugi, H. Ito, M. Nishimura, and K. H. Nagai, “C. elegans collectively forms dynamical networks,” Nature Communications, Vol.10, p. 683, 2019.
- [6] R. A. Cloney and E. Florey, “Ultrastructure of cephalopod chromatophore organs,” Zeitschrift für Zellforschung, Vol.89, pp. 250-280, 1968.
- [7] E. Florey, “Nervous control and spontaneous activity of the chromatophores of a cephalopod, Loligo opalescens,” Comp. Biochem. Physiol., Vol.18, Issue 2, pp. 305-324, 1966.
- [8] R. Sho, T. Yamagawa, H. Moribe, H. Ito, and K. Nakajima, “Speech recognition by chromatophores of squid via reservoir computing,” Proc. of 2021 Int. Symp. on Micro-NanoMechatronics and Human Science (MHS), 2021.
 This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.
This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.