Paper:
Computer-Based Blood Type Identification Using Image Processing and Machine Learning Algorithm
Marife A. Rosales† and Robert G. de Luna
Department of Electronics and Communications Engineering, De La Salle Lipa
1962 J.P. Laurel National Highway, Mataas na Lupa, Batangas 4217, Philippines
†Corresponding author
Blood type identification is a method used for determining the specific blood type of a person. It is a requirement before blood transfusions or blood donations is undertaken especially during emergency situations. Presently, the tests are performed manually by medical technologists in the laboratories. Sometimes, manual blood typing is prone to human error, resulting to incorrect blood grouping and wrong typing in the report, leading to fatal transfusion reactions. The study was focused on the development of a device that is capable of identifying the blood type of an individual using an image processing and machine learning algorithms. The study covered the identification of eight blood types, specifically rhesus positive and negative, A, B, O, and AB, by developing a capturing box integrated with a web camera system that could effectively capture blood sample images. In this study, the methodologies utilized were image processing through segmentation, feature extraction by color and texture properties, and different machine learning algorithms. After training, the results showed that coarse tree DT has the best performance accuracy score of 97.77% using 70:30 holdout validation. The testing results showed that the system is 100% accurate as validated by a registered medical technologist.
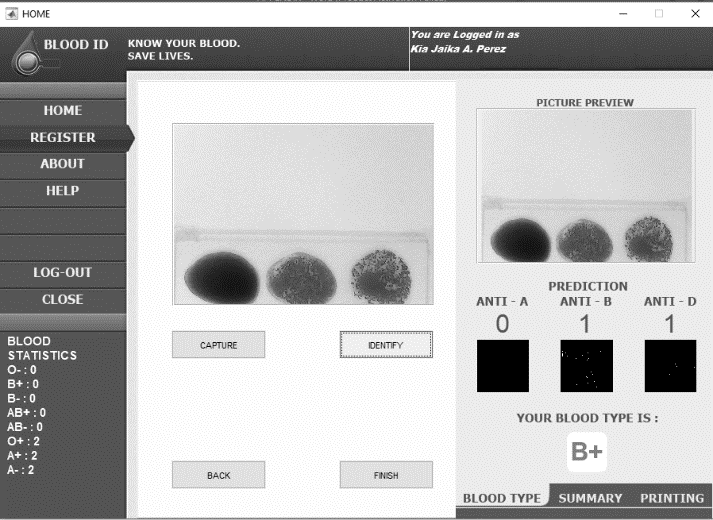
Blood type identification GUI with blood sample and blood type
- [1] A. Sahastrabuddhe and S. Ajij, “Blood Group Detection and RBC, WBC Counting: An Image Processing Approach,” Int. J. Eng. Comput. Sci., Vol.5, Issue 10, pp. 18635-18639, 2016.
- [2] M. Sidhu et al., “Report on errors in pretransfusion testing from a tertiary care center: A step toward transfusion safety,” Asian J. Transfus. Sci., Vol.10, No.1, pp. 48-52, 2016.
- [3] R. A. Rathod and R. A. Pathan, “Determination and Classification of Human Blood Types Using SIFT Transform and SVM Classifier,” Int. J. of Advanced Research in Electrical, Electronics and Instrumentation Engineering, Vol.5, Issue 11, pp. 8467-8473, 2016.
- [4] S. M. N. Fathima, “Classification of Blood Types by Microscope Color Images,” Int. J. Mach. Learn. Comput., Vol.3, No.4, pp. 376-379, 2013.
- [5] A. Ferraz, V. Carvalho, and F. Soares, “A Prototype for Blood Typing Based on Image Processing,” Sensordevices 4th Int. Conf. Sens. Device Technol. Appl., pp. 139-144, 2013.
- [6] G. Ravindran et al., “Determination and Classification of Blood Types Using Image Processing Techniques,” Int. J. Comput. Appl., Vol.157, No.1, pp. 12-16, 2017.
- [7] M. A. Rosales et al., “Faster R-CNN Based Fish Detector for Smart Aquaculture System,” IEEE 13th Int. Conf. Humanoid, Nanotechnology, Inf. Technol. Commun. Control. Environ. Manag., doi: 10.1109/HNICEM54116.2021.9732042, 2022.
- [8] S. S. Nikam, “A Comparative Study of Classification Techniques in Data Mining Algorithms,” Orient. J. Comput. Sci. Technol., Vol.8, No.1, pp. 13-19, 2015.
- [9] M. A. Rosales et al., “Physiological-Based Smart Stress Detector Using Machine Learning Algorithms,” IEEE 11th Int. Conf. Humanoid, Nanotechnology, Inf. Technol. Commun. Control. Environ. Manag. (HNICEM), doi: 10.1109/HNICEM48295.2019.9073355, 2019.
- [10] M. A. Rosales et al., “Artificial Intelligence: The Technology Adoption and Impact in the Philippines,” IEEE 12th Int. Conf. Humanoid, Nanotechnology, Inf. Technol. Commun. Control. Environ. Manag. (HNICEM), doi: 10.1109/HNICEM51456.2020.9400025, 2020.
- [11] S. Uchida, “Image processing and recognition for biological images,” Development, Growth & Differentiation, Vol.55, No.4, pp. 523-549, 2013.
- [12] J. M. Sharif et al., “Red blood cell segmentation using masking and watershed algorithm: A preliminary study,” Int. Conf. Biomed. Eng. (ICoBE), pp. 258-262, 2012.
- [13] K. B. Soni and S. S. Lokhande, “Blood Group Detection and WBC, RBC Cells Count,” Int. J. Innov. Res. Electr. Electron. Instrum. Control Eng., Vol.6, No.6, pp. 78-83, 2018.
- [14] N. M. D. Costa, “Automatic blood typing scanner through agglutination,” Ph.D. thesis, Universidade do Porto, 2014.
- [15] G. Mandloi, “A Survey on Feature Extraction Techniques for Color Images,” Int. J. Comput. Sci. Inf. Technol., Vol.3, No.3, pp. 4615-4620, 2013.
- [16] M. Hasantalukder et al., “Improvement of Accuracy of Human Blood Groups Determination Using Image processing Techniques,” Int. J. Adv. Res. Comput. Commun. Eng., Vol.4, No.10, pp. 411-412, 2015.
- [17] W. Zhu, N. Zeng, and N. Wang, “Sensitivity, Specificity, Accuracy, Associated Confidence Interval and ROC Analysis with Practical SAStextsuperscript® Implementations K&L Consulting Services, Inc., Fort Washington, PA Octagon Research Solutions, Wayne,” NESUG 2010 Heal. Care Life Sci., 2010. https://www.lexjansen.com/nesug/nesug10/hl/hl07.pdf [accessed June 12, 2019]
- [18] M. A. Rosales et al., “Non-Invasive Glycosylated Hemoglobin Monitoring Using Artificial Neural Network and Optimized SVM,” 9th Int. Symp. Comput. Intell. Ind. Appl., doi: 10.13140/RG.2.2.28403.45604, 2020.
- [19] D. Khanna et al., “Comparative Study of Classification Techniques (SVM, Logistic Regression and Neural Networks) to Predict the Prevalence of Heart Disease,” Int. J. Mach. Learn. Comput., Vol.5, No.5, pp. 414-419, 2015.
 This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.
This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.