Research Paper:
Noninvasive Optical Flow Analysis of White Blood Cell Dynamics for Enhanced COVID-19 Screening
Emi Yuda*,**,†
 , Yutaka Yoshida*, Itaru Kaneko*, Daisuke Hirahara*, and Junichiro Hayano***
, Yutaka Yoshida*, Itaru Kaneko*, Daisuke Hirahara*, and Junichiro Hayano***
*Innovation Center for Semiconductor and Digital Future (ICSDF), Mie University
1577 Kurima-Machiyacho, Tsu, Mie 514-8507, Japan
**Center for Data-Driven Science and Artificial Intelligence, Tohoku University
41 Kawauchi, Aoba-ku, Sendai, Miyagi 980-8576, Japan
***Nagoya City University
1 Kawasumi, Mizuho-cho, Mizuho-ku, Nagoya 467-8601, Japan
†Corresponding author
The COVID-19 pandemic has underscored the urgent need for enhanced first-line screening methods to complement traditional temperature checks and interviews prior to confirmatory polymerase chain reaction or rapid antigen testing. This study investigates the utility of white blood cell (WBC) counts as a predictive biomarker for COVID-19 identification and explores a novel, noninvasive approach for estimating WBC counts. Two key experiments were conducted. First, the predictive power of WBC counts was evaluated for COVID-19 detection using machine learning algorithms on a publicly available dataset. Second, a noninvasive optical flow analysis technique was proposed to estimate WBC counts from capillary blood flow images. The findings revealed that WBC was consistently selected as a significant feature across various feature selection methods. A LinearModel_BAG_L1 algorithm implemented with AutoGluon achieved an area under the receiver operating characteristic curve of 0.81 for COVID-19 prediction. Furthermore, the optical flow analysis method demonstrated a strong positive correlation (r=0.66) with conventional blood tests in estimating WBC counts. Although WBC counts alone may not provide sufficient diagnostic accuracy, the results highlight their value as a supplementary biomarker for preliminary COVID-19 screening. Additionally, the feasibility of noninvasive WBC estimation suggests promising applications in enhancing current testing frameworks and increasing accessibility to early detection tools.
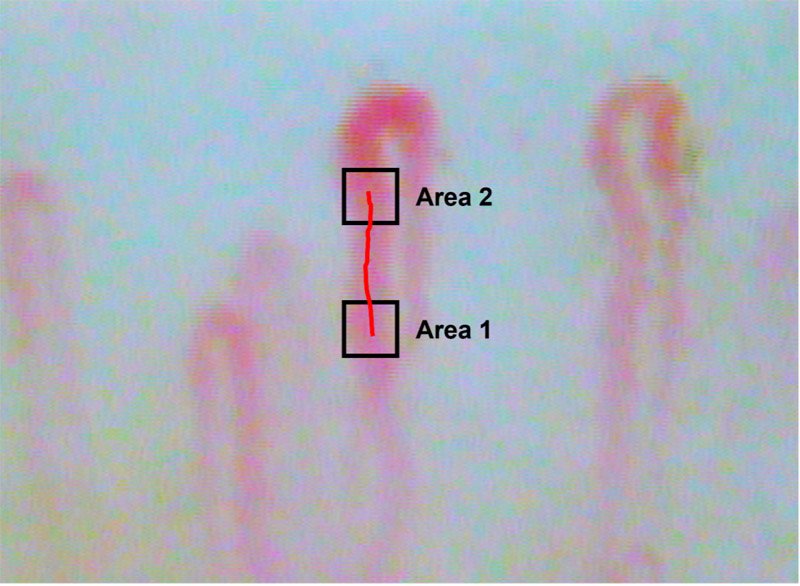
Observed white blood cell flow capillaries
- [1] C. L. Murrant and N. M. Fletcher, “Capillary communication: The role of capillaries in sensing the tissue environment, coordinating the microvascular, and controlling blood flow,” American J. of Physiology – Heart and Circulatory Physiology, Vol.323, Issue 5, pp. H1019-H1036, 2022. https://doi.org/10.1152/ajpheart.00088.2022
- [2] A. G. Hudetz, “Blood Flow in the Cerebral Capillary Network: A Review Emphasizing Observations with Intravital Microscopy,” Microcirculation, Vol.4, Issue 2, pp. 233-252, 1997. https://doi.org/10.3109/10739689709146787
- [3] J. Prothero and A. C. Burton, “The Physics of Blood Flow in Capillaries: I. The Nature of the Motion,” Biophysical J., Vol.1, Issue 7, pp. 565-579, 1961. https://doi.org/10.1016/S0006-3495(61)86909-9
- [4] H. N. T. Phuong, H. Jeong, and C. Shin, “Study on Image Processing of Capillaries Using Microscope: Initial Considerations,” Proc. of the 27th Int. Workshop on Frontiers of Computer Vision, pp. 157-167, 2021. https://doi.org/10.1007/978-3-030-81638-4_12
- [5] H. N. T. Phuong, H. Jeong, and C. Shin, “Consideration of Convolutional Neural Networks for Image Processing of Capillaries,” Proc. of the 2021 Int. Conf. on Artificial Intelligence in Information and Communication (ICAIIC), 2021. https://doi.org/10.1109/ICAIIC51459.2021.9415270
- [6] E. E. Manuelidis and L. Thomas, “Occlusion of brain capillaries by endothelial swelling in mycoplasma infections,” Proc. of the National Academy of Sciences of the United States of America, Vol.70, No.3, pp. 706-709, 1973. https://doi.org/10.1073/pnas.70.3.706
- [7] M. Palladino, “Complete blood count alterations in COVID-19 patients: A narrative review,” Biochemia Medica, Vol.31, Issue 3, Article No.030501, 2021. https://doi.org/10.11613/BM.2021.030501
- [8] C.-W. Hsu, J.-L. Lin, D.-T. Lin-Tan, T.-H. Yen, and K.-H. Chen, “White Blood Cell Count Predicts All-Cause, Cardiovascular Disease-Cause and Infection-Cause One-Year Mortality of Maintenance Hemodialysis Patients,” Therapeutic Apheresis and Dialysis, Vol.14, Issue 6, pp. 552-559, 2010. https://doi.org/10.1111/j.1744-9987.2010.00849.x
- [9] A. Jain and D. J. Doyle, “Apoptosis and pericyte loss in alveolar capillaries in COVID-19 infection: Choice of markers matters,” Intensive Care Medicine, Vol.46, pp. 1965-1966, 2020. https://doi.org/10.1007/s00134-020-06208-x
- [10] M. Drees, N. Kanapathippillai, and M. T. Zubrow, “Bandemia with Normal White Blood Cell Counts Associated with Infection,” American J. of Medicine, Vol.125, Issue 11, pp. 1124.E9-1124.E15, 2012. https://doi.org/10.1016/j.amjmed.2012.04.039
- [11] P. S. Aghbash, R. Rasizadeh, M. Shirvaliloo, J. S. Nahand, and H. B. Baghi, “Dynamic alterations in white blood cell counts and SARS-CoV-2 shedding in saliva: An infection predictor parameter,” Frontiers in Medicine, Vol.10, Article No.1208928, 2023. https://doi.org/10.3389/fmed.2023.1208928
- [12] Einstein Data4u, “Diagnosis of COVID-19 and its clinical spectrum.” https://www.kaggle.com/datasets/einsteindata4u/covid19 [Accessed September 16, 2023]
- [13] N. Compté, L. Dumont, D. Bron, S. De Breucker, J.-P. Praet, I. Bautmans, and T. Pepersack, “White blood cell counts in a geriatric hospitalized population: A poor diagnostic marker of infection,” Experimental Gerontology, Vol.114, pp. 87-92, 2018. https://doi.org/10.1016/j.exger.2018.11.002
- [14] Y. C. Fung, “Biomechanics: Circulation,” 2nd Edition, Springer, 1997. https://doi.org/10.1007/978-1-4757-2696-1
- [15] L. Wynants, B. Van Calster, G. S. Collins, R. D. Riley, G. Heinze, E. Schuit, E. Albu, B. Arshi, V. Bellou, M. M. J. Bonten, D. L. Dahly, J. A. Damen, T. P. A. Debray et al., “Prediction models for diagnosis and prognosis of COVID-19: Systematic review and critical appraisal,” BMJ, Vol.369, Article No.m1328, 2020. https://doi.org/10.1136/bmj.m1328
- [16] S. Kurstjens, A. van der Horst, R. Herpers, M. W. L. Geerits, Y. C. M. Kluiters-de Hingh, E.-L. Göttgens, M. J. T. Blaauw, M. H. M. Thelen, M. G. L. M. Elisen, and R. Kusters, “Rapid identification of SARS-CoV-2-infected patients at the emergency department using routine testing,” Clinical Chemistry and Laboratory Medicine, Vol.58, Issue 9, pp. 1587-1593, 2020. https://doi.org/10.1515/cclm-2020-0593
- [17] N. Chen, M. Zhou, X. Dong, J. Qu, F. Gong, Y. Han, Y. Qiu, J. Wang, Y. Liu, Y. Wei et al., “Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study,” The Lancet, Vol.395, Issue 10223, pp. 507-513, 2020. https://doi.org/10.1016/S0140-6736(20)30211-7
- [18] C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu et al., “Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China,” The Lancet, Vol.395, Issue 10223, pp. 497-506, 2020. https://doi.org/10.1016/S0140-6736(20)30183-5
- [19] L. Yan, H.-T. Zhang, J. Goncalves, Y. Xiao, M. Wang, Y. Guo, C. Sun, X. Tang, L. Jing, M. Zhang et al., “An interpretable mortality prediction model for COVID-19 patients,” Nature Machine Intelligence, Vol.2, pp. 283-288, 2020. https://doi.org/10.1038/s42256-020-0180-7
- [20] D. Wang, B. Hu, C. Hu, F. Zhu, X. Liu, J. Zhang, B. Wang, H. Xiang, Z. Cheng, Y. Xiong et al., “Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China,” JAMA, Vol.323, No.11, pp. 1061-1069, 2020. https://doi.org/10.1001/jama.2020.1585
- [21] C. Qin, L. Zhou, Z. Hu, S. Zhang, S. Yang, Y. Tao, C. Xie, K. Ma, K. Shang, W. Wang, and D.-S. Tian, “Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China,” Clinical Infectious Diseases, Vol.71, Issue 15, pp. 762-768, 2020. https://doi.org/10.1093/cid/ciaa248
- [22] L. Tan, Q. Wang, D. Zhang, J. Ding, Q. Huang, Y.-Q. Tang, Q. Wang, and H. Miao, “Lymphopenia predicts disease severity of COVID-19: A descriptive and predictive study,” Signal Transduction and Targeted Therapy, Vol.5, Article No.33, 2020. https://doi.org/10.1038/s41392-020-0148-4
- [23] J. Liu, Y. Liu, P. Xiang, L. Pu, H. Xiong, C. Li, M. Zhang, J. Tan, Y. Xu, R. Song et al., “Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage,” J. of Translational Medicine, Vol.18, Article No.206, 2020. https://doi.org/10.1186/s12967-020-02374-0
- [24] L. Wang, “C-reactive protein levels in the early stage of COVID-19,” Médecine et Maladies Infectieuses, Vol.50, Issue 4, pp. 332-334, 2020. https://doi.org/10.1016/j.medmal.2020.03.007
 This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.
This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.