Paper:
Robust Cell Image Segmentation via Improved Markov Random Field Based on a Chinese Restaurant Process Model
Dongming Li*1,*2, Changming Sun*3, Su Wei*4, Yue Yu*2,*5, and Jinhua Yang*1,†
*1College of Opto-Electronic Engineering, Changchun University of Science and Technology
No.7089 Weixin Road, Chaoyang District, Changchun, Jilin 130022, China
*2School of Information Technology, Jilin Agricultural University
No.2888 Xincheng Road, Jingyue District, Changchun, Jilin 130118, China
*3CSIRO Data61
P.O. Box 76, Epping, New South Wales 1710, Australia
*4Modern Educational Technology Center, Changchun University of Chinese Medicine
No.1035 Boshuo Road, Jingyue District, Changchun, Jilin 130117, China
*5College of Artificial Intelligence, Tourism College of Changchun University
Sheling Town, University Campus District, Changchun, Jilin 130607, China
†Corresponding author
In this paper, a segmentation method for cell images using Markov random field (MRF) based on a Chinese restaurant process model (CRPM) is proposed. Firstly, we carry out the preprocessing on the cell images, and then we focus on cell image segmentation using MRF based on a CRPM under a maximum a posteriori (MAP) criterion. The CRPM can be used to estimate the number of clusters in advance, adjusting the number of clusters automatically according to the size of the data. Finally, the conditional iteration mode (CIM) method is used to implement the MRF based cell image segmentation process. To validate our proposed method, segmentation experiments are performed on oral mucosal cell images. The segmentation results were compared with other methods, using precision, Dice, and mean square error (MSE) as the objective evaluation criteria. The experimental results show that our method produces accurate cell image segmentation results, and our method can effectively improve segmentation for the nucleus, binuclear cell, and micronucleus cell. This work will play an important role in cell image recognition and analysis.
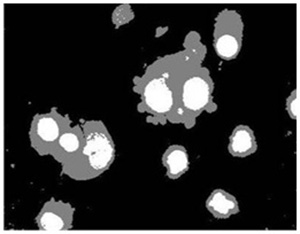
Cell image segmentaion by our method
- [1] A. Gharipour and A. W.-C Liew, “Segmentation of cell nuclei in fluorescence microscopy images: An integrated framework using level set segmentation and touching-cell splitting,” Pattern Recognition, Vol.58, pp. 1-11, 2016.
- [2] J. C. Dunn, “A Fuzzy Relative of the ISODATA Process and Its Use in Detecting Compact Well-Separated Clusters,” J. of Cybernetics, Vol.3, pp. 32-57, 1973.
- [3] H. P. Ng, S. H. Ong, K. W. C. Foong et al., “Medical Image Segmentation Using K-Means Clustering and Improved Watershed Algorithm,” IEEE Southwest Symp. on Image Analysis and Interpretation, pp. 61-65, 2006.
- [4] X. Zong and W. Tian, “Segmentation and feature extraction of brain tumor based on magnetic resonance image using k-means,” Computer Engineering and Applications, Vol.1, pp. 1-10, 2019.
- [5] Q. Tuo, “Research on Image Threshold Segmentation Algorithm based on Maximum Entropy and Genetic Algorithm,” Kunming University of Science and Technology, pp. 20-31, 2016 (in Chinese).
- [6] Y. Meng, G. Liu, S. Xu, and F. Feng, “Image Segmentation Method Using Multi-Resolution Markov Random Field Model with Edge-Preserving,” J. of Xi’an Jiaotong University, Vol.53, pp. 56-65, 2019.
- [7] R. Millioni, S. Sbrignadello, A. Tura, E. Iori, E. Murphy, and P. Tessari, “The inter- and intra-operator variability in manual spot segmentation and its effect on spot quantitation in two-dimensional electrophoresis analysis,” Electrophoresis, Vol.31, No.10, pp. 1739-1742, 2010.
- [8] J. E. Iglesias, “Globally optimal coupled surfaces for semi-automatic segmentation of medical images,” Information Processing in Medical Imaging: Int. Conf. on Information Processing in Medical Imaging, Vol.10265, pp. 610-621, 2017.
- [9] R. D. Stewart, I. Fermin, and M. Opper, “Region growing with pulse-coupled neural networks: an alternative to seeded region growing,” IEEE Trans. on Neural Networks, Vol.13, No.6, pp. 1557-1562, 2002.
- [10] H. Zhou, Z. Wang, W. Lin, and B. Ding, “An Improved Segmentation Method for Cytoskeleton Images,” Pattern Recognition and Artificial Intelligence, Vol.21, No.2, pp. 269-274, Apr. 2008.
- [11] N. Otsu, “A threshold selection method from gray-level histograms,” IEEE Trans. on System, Man, and Cybernetics, Vol.9, No.1, pp. 62-66, 1979.
- [12] T. F. Chan and L. A. Vese, “Active contours without edges,” IEEE Trans. on Image Processing, Vol.10, No.2, pp. 266-277, 2001.
- [13] H.-Y. Jiang, Y.-P. Si, and X.-G. Luo, “Medical Image Segmentation Based on Improved Ostu Algorithm and Regional Growth Algorithm,” J. of Northeastern University, Natural Science, Vol.27, No.4, pp. 398-401, 2006.
- [14] J. A. Sethian, “A fast marching level set method for monotonically advancing fronts,” Proc. of the National Academy of Sciences of the United States of America, Vol.93, No.4, pp. 1591-1595, 1996.
- [15] C. Li, C. Xu, C. Gui et al., “Level set evolution without re-initialization: a new variational formulation,” Proc. of the 2005 IEEE Computer Society Conf. on Computer Vision and Pattern Recognition, Vol.1, pp. 430-436, 2005.
- [16] S. Lankton and A. Tannenbaum, “Localizing region-based active contours,” IEEE Trans. Image Process, Vol.17, No.11, pp. 2029-2039, 2008.
- [17] J. Kaur and A. Jindal, “Comparison of thyroid segmentation algorithms in ultrasound and scintigraphy images,” Int. J. of Computer Applications, Vol.50, No.23, pp. 24-27, 2012.
- [18] T. Kanungo, D. M. Mount, N. S. Netanyahu, C. D. Piatko, R. Silverman, and A. Y. Wu, “An efficient k-means clustering algorithm: analysis and implementation,” IEEE Trans. on Pattern Analysis and Machine Intelligence, Vol.24, No.7, pp. 881-892, 2002.
- [19] S. A. Mingoti and J. O. Lima, “Comparing SOM neural network with fuzzy c-means, K-means and traditional hierarchical clustering algorithms,” European J. of Operational Research, Vol.174, No.3, pp. 1742-1759, 2006.
- [20] J. Long, E. Shelhamer, and T. Darrell, “Fully convolutional networks for semantic segmentation,” Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), pp. 3431-3440, 2015.
- [21] O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional networks for biomedical image segmentation,” Int. Conf. on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2015), Vol.9351, pp. 234-241, 2015.
- [22] Z. Jia, X. Huang, E. I.-C. Chang, and Y. Xu, “Constrained Deep Weak Supervision for Histopathology Image Segmentation,” IEEE Trans. on Medical Imaging, Vol.36, No.11, pp. 2376-2388, 2017.
- [23] D. Li, L. Zhang, C. Sun, T. Yin, C. Liu, and J. Yang, “Robust Retinal Image Enhancement via Dual-Tree Complex Wavelet Transform and Morphology-Based Method,” IEEE Access, Vol.7, pp. 47303-47316, 2019.
- [24] D.-M. Li, L.-J. Zhang, J.-H. Yang, and W. Su, “Research on wavelet-based contourlet transform algorithm for adaptive optics image denoising,” Optik, Vol.127, No.12, pp. 5029-5034, 2016.
- [25] S. Geman and D. Geman, “Stochastic Relaxation, Gibbs Distributions, and the Bayesian Restoration of Images,” IEEE Trans. on Pattern Analysis and Machine Intelligence, Vol.PAMI-6, No.6, pp. 721-741, 1984.
- [26] A. Andalib and S. M. Babamir, “A class-based link prediction using distance dependent Chinese restaurant process,” Physica A: Statistical Mechanics and Its Applications, Vol.456, pp. 204-214, 2016.
- [27] J.-L. Liu, Q. Sui, and W.-X. Zhu, “MR image segmentation based on probability density function and active contour model,” Optic and Precision Engineering, Vol.22, No.12, pp. 3435-3443, 2014.
 This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.
This article is published under a Creative Commons Attribution-NoDerivatives 4.0 Internationa License.